| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website https://jh.elmerpub.com |
Review
Volume 14, Number 2, April 2025, pages 43-55
MicroRNA Signatures: Illuminating Minimal Residual Disease Monitoring in Juvenile Myelomonocytic Leukemia - A Review
Bhavyadharshini Aruna, b, Geofrey Johnb, c, Rajeshkumar Ramand, e
aHasan Lab, Department of Medical Oncology, Advanced Centre for Treatment Research and Education in Cancer, Tata Memorial Centre, Navi Mumbai, Maharashtra, India
bHomi Bhabha National Institute, Mumbai, Maharashtra, India
cDepartment of Radiation Oncology, Advanced Centre for Treatment Research and Education in Cancer, Tata Memorial Centre, Navi Mumbai, Maharashtra, India
dDepartment of Pharmaceutical Biotechnology, JSS College of Pharmacy, Ooty, The Nilgiris, India
eCorresponding Author: Rajeshkumar Raman, Department of Pharmaceutical Biotechnology, JSS College of Pharmacy, Rocklands, Post Box No. 20, Udhagamandalam 643 001, The Nilgiris, India
Manuscript submitted November 22, 2024, accepted January 20, 2025, published online April 25, 2025
Short title: MRD Monitoring in JMML
doi: https://doi.org/10.14740/jh1384
- Abstract
- Introduction
- MiRNAs
- MRD Detection Techniques
- Comparison With Traditional MRD Monitoring Methods
- Impact of MiRNA Dysregulation on Disease Progression
- Clinical Applications
- Challenges and Future Directions
- Ethical Considerations
- Conclusion
- References
| Abstract | ▴Top |
Juvenile myelomonocytic leukemia (JMML) is an aggressive pediatric myelodysplastic/myeloproliferative neoplasm characterized by RAS pathway mutations and significant heterogeneity. Minimal residual disease (MRD) monitoring is crucial for evaluating treatment response and predicting relapse risk. MicroRNA (miRNAs), small non-coding RNAs with pivotal roles in gene regulation, have emerged as promising biomarkers for JMML MRD detection. This review explores the mechanistic role of miRNAs in JMML pathogenesis, emphasizing their diagnostic, prognostic, and therapeutic potential. Dysregulated miRNA profiles correlate with distinct JMML subgroups and disease progression, suggesting utility in non-invasive MRD monitoring. Emerging evidence highlights miR-150-5p as a tumor suppressor targeting STAT5b and its therapeutic potential in ameliorating JMML’s aberrant signaling pathways. We compare traditional MRD methods, such as flow cytometry and polymerase chain reaction (PCR), with miRNA-based techniques, underscoring the latter’s superior sensitivity, specificity, and non-invasiveness. Recent advances in miRNA profiling technologies, including next-generation sequencing and digital PCR, enable precise detection of residual leukemic cells and support personalized treatment approaches. Despite significant progress, challenges persist in standardizing miRNA-based assays and validating their clinical utility. Ethical considerations, including patient privacy and informed consent, remain critical for integrating miRNA diagnostics into routine care. This review provides a comprehensive synthesis of current knowledge on miRNA signatures in JMML, illuminating their transformative potential in MRD monitoring and paving the way for innovative therapeutic strategies.
Keywords: MicroRNA; Minimal residual disease; Juvenile myelomonocytic leukemia; MRD detection techniques; MiRNA profiling
| Introduction | ▴Top |
Juvenile myelomonocytic leukemia (JMML) is an uncommon hematological malignancy affecting young children, predominantly under the age of 6, with varying degrees of clinical aggressiveness depending on the subtype. Classified by the World Health Organization (WHO) as a myeloproliferative neoplasm (MPN), JMML is characterized by a clonal proliferation of abnormal myeloid cells within the bone marrow, peripheral blood, and other tissues [1-3] (Fig. 1).
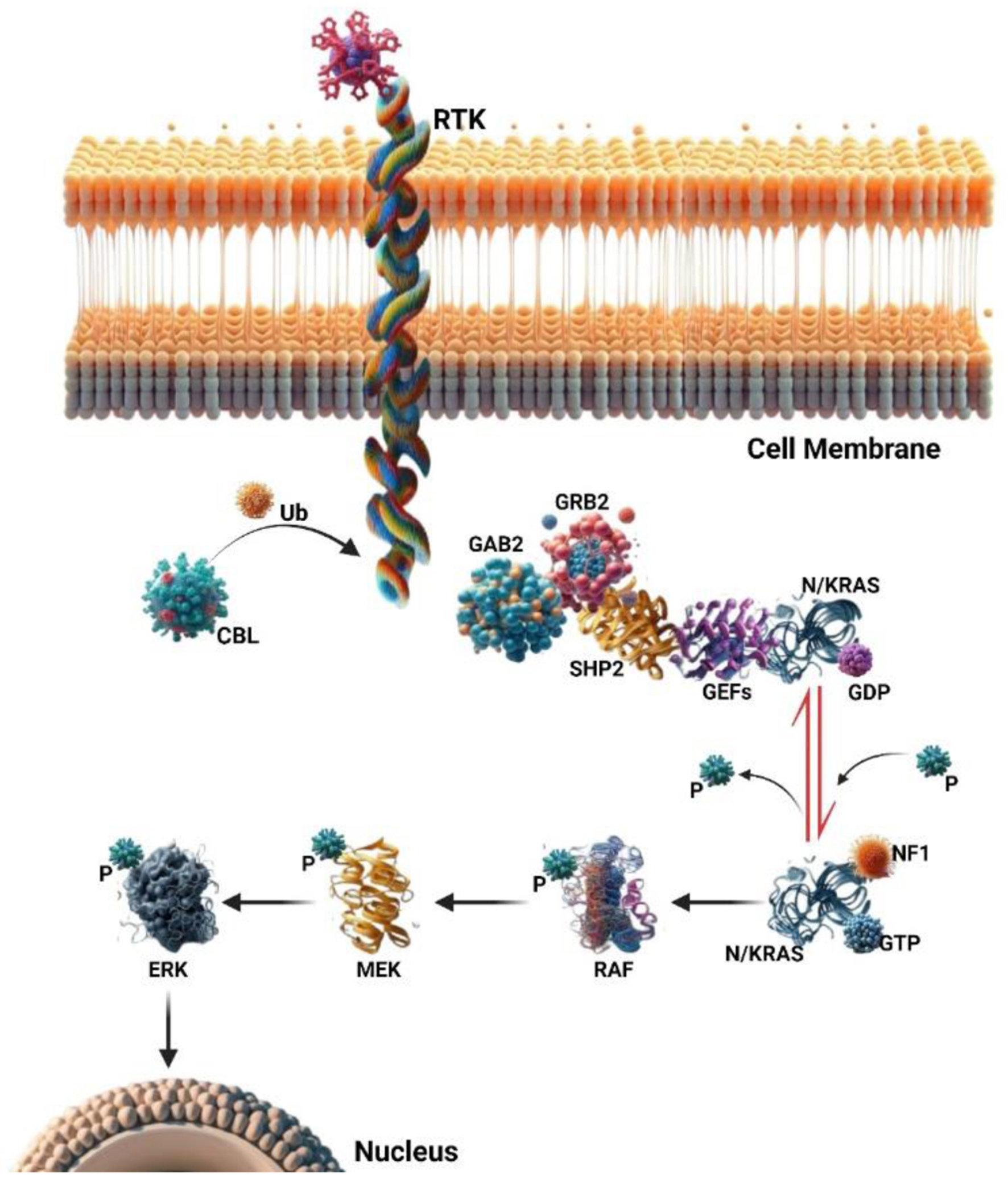 Click for large image | Figure 1. The RAS signaling pathway involves small GTPase switch proteins, NRAS, and KRAS. These proteins act downstream of receptor and non-receptor tyrosine kinases (RTKs and TKs). RAS activation status is regulated through phosphorylation and dephosphorylation changes. Guanine nucleotide-exchange factors (GEFs) and GTPase activating proteins (GAPs) play opposing roles in this regulation. Upon RTK or TK stimulation, adaptor proteins (such as GAB2, GRB2, and SHP2) recruit GEFs, leading to RAS-GDP phosphorylation and activation of RAS-GTP. Active RAS then triggers a signaling cascade, sequentially activating RAF, MEK, and ERK proteins. These downstream signals ultimately control cell functions like proliferation, survival, and differentiation. GAPs (e.g., NF1) inactivate the RAS pathway by promoting RAS-GTP dephosphorylation to an inactive RAS-GDP form. Additionally, the ubiquitin ligase CBL negatively regulates the RAS pathway by targeting active RTKs for proteasomal degradation. |
Secondary mutations, including those in transcription factors like RUNX1 and GATA2, and spliceosome components such as ZRSR2, further exacerbate disease progression and are associated with higher relapse rates and poorer outcomes. This genetic complexity is reflected in JMML’s clinical presentation, which often includes splenomegaly, hepatomegaly, anemia, thrombocytopenia, and systemic symptoms such as fever and weight loss. Autoimmune manifestations, such as vasculitis and arthritis, are observed in a subset of patients with JMML, particularly those with mutations in the RAS pathway (e.g., PTPN11, NRAS, KRAS, NF1, and CBL) [4, 5]. These manifestations can precede formal diagnosis, indicating an underlying autoimmune process linked to the disease [6]. For instance, a case report detailed a 7-year-old boy with JMML who developed severe steroid-dependent uveitis, an inflammatory eye condition [7]. Such diverse clinical features, combined with JMML’s rarity, complicate its diagnosis and management, often delaying treatment initiation [8, 9].
The current standard of care for JMML is hematopoietic stem cell transplantation (HSCT), which offers the only potential for long-term disease control. However, relapse rates remain significant, with overall survival ranging between 52% and 63% [10]. Allogeneic HSCT is currently the only curative treatment, but it is associated with significant challenges. Studies have reported 3-year overall survival (OS) rates of approximately 63% and event-free survival (EFS) rates around 52%. Additionally, transplantation-related mortality (TRM) contributes to the overall mortality, with some studies noting TRM rates of approximately 21% [11]. Advances in molecular biology have highlighted the potential of targeted therapies, including tyrosine kinase inhibitors (TKIs), mitogen-activated protein kinase (MEK) inhibitors, and mTOR inhibitors, as adjuncts to HSCT. These agents target the dysregulated signaling pathways driving JMML pathogenesis, such as the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR cascades. Despite their promise, these treatments remain experimental and necessitate robust biomarkers to refine patient stratification, monitor treatment response, and predict outcomes [12].
Minimal residual disease (MRD) monitoring has emerged as a cornerstone of JMML management, enabling the detection of residual leukemic cells that are undetectable by conventional diagnostic modalities [13]. MRD serves as a prognostic indicator for disease relapse and guides therapeutic decisions, including the intensification or de-escalation of treatment regimens. In JMML, MRD monitoring, typically using flow cytometry (FCM) and polymerase chain reaction (PCR), is essential but not yet standardized, unlike in other leukemias such as acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), or chronic lymphocytic leukemia (CLL). Recent advances, such as droplet digital PCR (ddPCR), show promise in detecting relapse earlier, particularly after HSCT [14]. While these methods are effective, they have limitations, including invasiveness, moderate sensitivity, and dependence on bone marrow aspirates. These challenges highlight the need for novel, non-invasive, and more sensitive approach to MRD monitoring [15-17].
An important advancement is the incorporation of MRD evaluations into standard clinical decision-making protocols. Recent research has identified microRNAs (miRNAs) as promising biomarkers for MRD detection in JMML [18]. MiRNAs are small, non-coding RNA molecules that regulate gene expression post-transcriptionally by binding to target mRNAs, leading to their degradation or translational repression. Dysregulated miRNA expression has been implicated in various cancers, including hematological malignancies, where they play oncogenic or tumor-suppressive roles. In JMML, specific miRNA signatures have been associated with disease pathogenesis, prognosis, and therapeutic response. For instance, studies have demonstrated the tumor-suppressive role of miR-150-5p, which targets STAT5b, a regulator of myeloid proliferation. The downregulation of miR-150-5p in JMML correlates with enhanced leukemic cell growth and poor clinical outcomes [19].
Advancements in molecular profiling technologies, such as next-generation sequencing (NGS) and single-cell RNA sequencing, have significantly enhanced the precision of miRNA detection and characterization. These techniques enable the identification of miRNA expression patterns with unprecedented sensitivity and specificity, allowing for earlier disease detection and better relapse prediction. Additionally, miRNA profiling can be performed using easily accessible biological samples, such as blood or serum, reducing the need for invasive procedures like bone marrow biopsies. This non-invasive approach is particularly advantageous in pediatric patients, minimizing discomfort and associated risks.
The integration of miRNA-based MRD monitoring into JMML management holds great promise for improving patient outcomes. While miRNA profiling has shown promise in detecting neoplastic miRNAs in various cancers, its clinical utility in JMML remains limited. Studies have identified specific miRNAs, such as miR-150-5p, as potential tumor suppressors in JMML as previously discussed [19]. However, these findings have not yet translated into clinically actionable results for disease monitoring or personalized treatment strategies. In contrast, miRNA profiling has demonstrated more established clinical applications in other cancers. For instance, in prostate cancer, miRNAs have been utilized as biomarkers for diagnosis, prognosis, and therapy response assessment [20, 21]. Similarly, in colorectal cancer, deregulated miRNAs have been associated with disease progression, highlighting their potential as diagnostic and prognostic tools [22].
Moreover, as miRNA research continues to evolve, new therapeutic opportunities may emerge, including miRNA mimics and inhibitors that target dysregulated pathways in JMML [23]. miR-126 is upregulated in leukemic stem cells (LSCs) in AML, where its overexpression promotes cell quiescence and reduces differentiation, contributing to chemotherapy resistance [24]. Targeting miR-126 with antagomiRs delivered via lipid nanoparticles has shown promising results, reducing AML blasts by 80% in vitro and improving survival in mouse models [25]. It may also enhance the efficacy of existing chemotherapies. MiR-9, involved in the transformation of hematopoietic progenitor cells, promotes monocytic differentiation and has therapeutic potential by targeting the LIN28B/Let-7/HMGA2 axis [26]. Other miRNAs like miR-29b, miR-181a, and miR-155 are also being explored for their ability to inhibit leukemic cell growth and improve treatment outcomes in AML, with miR-155 currently in clinical trials [27].
Despite these advances, challenges remain in the standardization of miRNA-based assays and their validation in clinical settings. Future research should focus on addressing these challenges to facilitate the translation of miRNA biomarkers into routine clinical practice [15].
The discovery and application of miRNA signatures in JMML represent a significant step forward in the field of pediatric oncology. These biomarkers not only provide a powerful tool for MRD monitoring but also offer new insights into JMML pathogenesis and therapeutic vulnerabilities. As research progresses, the integration of miRNA-based approaches with existing diagnostic and therapeutic modalities has the potential to transform JMML management, improving outcomes for affected children worldwide.
| MiRNAs | ▴Top |
Biogenesis and function
MiRNAs are short regulatory non-coding RNAs, typically composed of 19-25 nucleotides in length [28]. MiRNAs, integral RNA regulators of gene expression, have emerged as a central subject of investigation in the context of molecular biology [29]. They constitute a family of molecules crucial for modulating the types and quantities of proteins synthesized within cells, thereby playing a pivotal role in the regulation of gene expression post-transcriptionally by binding to target mRNAs, resulting in gene silencing. These miRNA molecules are ubiquitously present within cells as well as in the circulatory system [30]. They are also found in various genomic locations including, intragenic regions introns of non-coding and coding genes, and exons of non-coding genes. Research has shown that miRNAs regulate numerous cellular functions such as cell growth, differentiation proliferation, and apoptosis. They are also involved in specifying tissue identity during differentiation processes, making them valuable markers for identifying specific cell types [31].
MiRNAs are transcribed as primary miRNA (pri-miRNA) molecules from precursor genes (mir-genes) by RNA polymerase II in the nucleus. These transcripts undergo splicing and capping, forming double-stranded, stem-loop structures with 5′ capping and 3′ polyadenylation. Pri-miRNAs are processed by Drosha, a nuclear RNase III enzyme, in collaboration with its cofactor DGCR8. Drosha cleaves the stem-loop structure, releasing precursor miRNA (pre-miRNA), which is transported to the cytoplasm by Exportin 5 and Ran-GTP. In the cytoplasm, the RNase III enzyme Dicer further processes pre-miRNAs by cleaving the double-stranded region to generate mature miRNA duplexes. This step involves the recognition of 2-nucleotide 3′ overhangs by Dicer’s PAZ domain. The mature miRNA duplex is then loaded onto the RNA-induced silencing complex (RISC), with Argonaute (Ago) proteins, particularly Ago2, playing a central role. Ago2 cleaves the passenger strand, leaving the guide strand to bind target mRNAs. The level of complementarity between the miRNA and its target determines the outcome: partial binding inhibits translation, while perfect binding leads to mRNA degradation [28, 31] (Fig. 2).
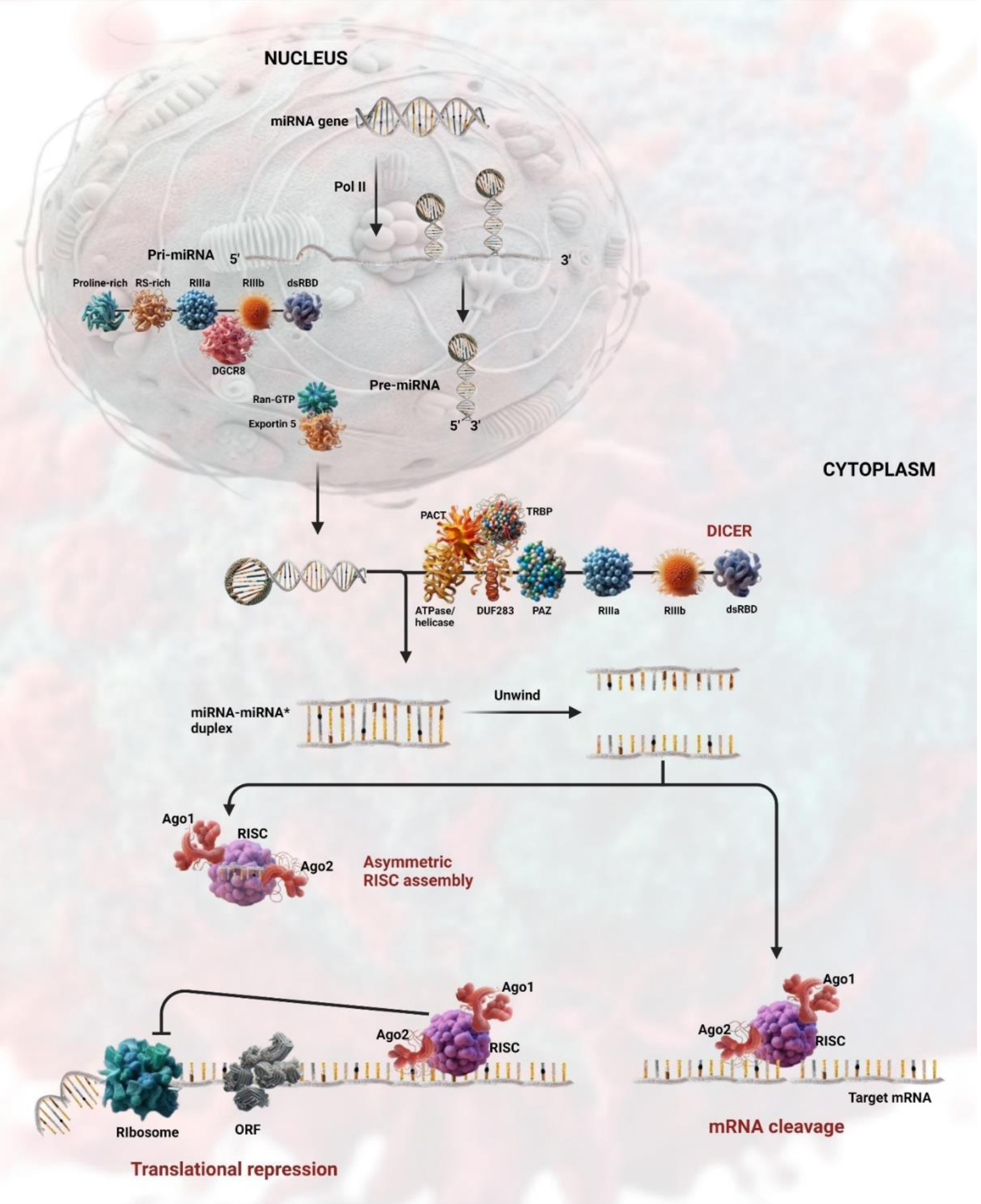 Click for large image | Figure 2. Schematic diagram of microRNA (miRNA) biogenesis and function pathway. |
Alternative pathways for miRNA biogenesis include mirtron formation by spliceosomes and the processing of short hairpin RNAs (shRNAs) into miRNAs. These mechanisms, along with their interaction with RISC, enable miRNAs to regulate gene expression post-transcriptionally, highlighting their versatility and functional significance in cellular processes [32].
Crosstalk between miRNAs and signalling pathways
These small non-coding RNAs, which regulate gene expression, have been found to play a crucial role in the progression of various diseases, influencing numerous aspects of cellular function. For instance, miRNAs are involved in modulating neuroinflammation in Parkinson’s disease by targeting inflammatory pathways, such as miR-146a’s regulation of Toll-like receptor 4 (TLR4) and nuclear factor (NF)-kappaB signaling, which mitigates inflammation [33]. In cancer, miRNAs act as oncogenes or tumor suppressors, influencing processes like cell proliferation and metastasis through mechanisms such as regulating E2F transcription factors and epithelial-mesenchymal transition [34]. Alterations in miRNA expression levels across various diseases underscore their significance in pathogenesis and their potential as biomarkers and therapeutic targets [35]. One of the most extensively studied interactions is between miRNAs and the Wnt/β-catenin signaling pathway. This pathway plays a key role in cellular processes, and a study by Peng et al suggested that specific miRNAs can modulate its activity. This modulation can impact various cellular processes, adding another layer of complexity to our understanding of disease progression [36].
But the story does not end there. Another fascinating aspect of miRNA research is the interaction between miRNAs and circular RNAs (circRNAs). Recent research has increasingly focused on the interaction between miRNA and circRNAs, highlighting their significant roles in gene regulation and cellular processes. For example, a study utilizing a novel sequencing technology identified hundreds of mRNA-interacting circRNAs, suggesting that these interactions play a crucial role in gene expression regulation [37]. Additionally, circRNAs have been shown to act as miRNA sponges, influencing transcriptional and post-transcriptional regulation, which opens new avenues for understanding complex diseases [38]. This crosstalk between miRNAs and circRNAs is essential for comprehending the regulatory networks involved in disease progression, as evidenced by their involvement in various conditions such as cancer and diabetes [39, 40].
The interplay between Wnt-signaling pathways and miRNAs is particularly relevant in the context of cancer. Recent studies have demonstrated that a network of miRNAs influences Wnt-signaling, contributing to cancer development and progression. For instance, research has shown that miRNAs can activate or inhibit the Wnt/β-catenin pathway at various levels, with specific miRNAs targeting Wnt antagonists to enhance signaling, thereby promoting oncogenesis and metastasis [36, 41]. This highlights the potential of miRNA research in developing new therapeutic strategies for cancer treatment, as understanding these interactions could lead to targeted therapies that manipulate these signaling pathways effectively [42].
Role of miRNAs in hematological malignancies
In hematological malignancies, such as leukemias, lymphomas, and myeloma, which account for approximately 10-12% of annual cancer cases in the USA, miRNAs play a pivotal role in disease progression. Dysregulation of miRNAs in these malignancies often arises from genomic alterations, epigenetic modifications, or transcriptional disruptions, emphasizing their complex role in cancer pathology [43]. For instance, exosomal miRNAs from CLL cells promote proangiogenic and immunosuppressive factors within the tumor microenvironment [44]. Disruptions in miRNA biogenesis and post-transcriptional regulation are central to hematological cancers. Specific miRNAs like miR-155 are consistently overexpressed across multiple malignancies, while others, such as miR-150, show variable expression patterns [45]. In diffuse large B-cell lymphoma (DLBCL), the upregulation of miR-155, miR-221, and miR-21 correlates with prognosis [44]. Conversely, therapies like all-trans retinoic acid (ATRA) in acute promyelocytic leukemia (APL) have been shown to restore tumor-suppressive miRNAs.
MiRNA dysregulation in JMML
While substantial research has elucidated the role of miRNAs in leukemia, their specific involvement in JMML remains underexplored. Recent studies have highlighted significant findings regarding miRNA expression in JMML. For instance, elevated levels of miR-15 and miR-223 were observed in human-induced pluripotent stem cells (hiPSCs) derived from JMML patients with PTPN11 mutations, a common genetic alteration in this disease. This observation was validated in mononuclear cells from JMML patients, where inhibiting miR-223 in hiPSC-derived myeloid cells restored normal hematopoiesis, suggesting a critical role for this miRNA in disease pathology [46]. Additionally, the LIN28B gene’s overexpression was linked to increased hemoglobin F (HbF) levels and decreased expression of let-7 family miRNAs, correlating with poor clinical outcomes in JMML patients [47]. These findings indicate that dysregulated miRNA expression may contribute significantly to JMML pathogenesis. Specific miRNA expression signatures have also been identified that correlate with molecular subgroups of JMML, revealing distinct patterns of downregulated and upregulated miRNAs [48]. Moreover, certain miRNAs are associated with disease progression and survival rates, enabling risk stratification and the development of personalized treatment strategies. For example, miR-150-5p has been identified as a tumor suppressor in JMML. Its restoration may enhance sensitivity to therapeutic agents like granulocyte-macrophage colony-stimulating factor (GM-CSF), illustrating the therapeutic potential of targeting dysregulated miRNAs [19]. Furthermore, the identification of distinct miRNA fingerprints associated with specific molecular subtypes of JMML paves the way for more tailored interventions based on individual patient profiles. Ongoing research into these small RNA molecules not only enhances our understanding of JMML but also holds promise for improving diagnostic accuracy and therapeutic outcomes.
MiRNAs: pioneering tools in disease diagnosis, prognosis, and therapeutic advancement
MiRNAs play a critical role as diagnostic biomarkers, especially in cases where traditional methods fail to identify the disease. Their distinct expression profiles can serve as diagnostic biomarkers, especially in cases where traditional methods fail to provide clear insights. For example, specific miRNA signatures have been identified that can differentiate between ALL and other types of leukemia, enhancing diagnostic accuracy and enabling earlier intervention [49]. As prognostic biomarkers, miRNAs are linked to leukemia progression and treatment responses. They influence critical factors such as ATP-binding cassette (ABC) transporters, which play a significant role in drug resistance among ALL patients [50]. In B-cell acute lymphoblastic leukemia (B-ALL), miRNAs can also function as therapeutic targets by regulating protein expression through mRNA translation. Emerging technologies, such as miRNA mimics and inhibitors, show promise for therapeutic applications, although these strategies are still in the early stages of development [51]. The differential expression of miRNAs between pediatric and adult ALL underscores the urgent need for further research, particularly to address the less effective chemotherapy outcomes observed in adults [52]. Large-scale clinical trials are essential to validate miRNA-based diagnostics and therapies. Additionally, deeper insights into the interactions between miRNAs and immune cells could pave the way for improved immunotherapy strategies [53]. For instance, researchers have utilized data from the Chinese Glioma Genome Atlas (CGGA) to establish miRNA signatures that predict glioma patient survival, emphasizing the expanding role of miRNAs in precision medicine [54]. In JMML, where prognosis remains poor despite HSCT, miRNA research offers new insights. Studies have identified downregulated miRNAs such as miR-150-5p and let-7 that are linked to key signaling pathways like RAS and GM-CSF. MiR-150-5p acts as a tumor suppressor by targeting STAT5b; its restoration has been shown to reduce JMML cell proliferation and aberrant signaling [19]. These findings suggest that miRNA-based therapies could significantly improve outcomes for JMML patients, paving the way for future clinical applications.
| MRD Detection Techniques | ▴Top |
Conventional MRD detection methods
MRD detection through phenotyping identifies mutated leukemic progenitor cells in the bone marrow using multiparametric FCM. This technique compares post-relapse bone marrow samples with pretreatment leukemic cell immunophenotypes. Fluorescently tagged antibodies specific to leukemia markers facilitate MRD identification. Tracking B-cell maturation through FCM reveals marker changes between normal and leukemic pre-B-cell phenotypes. Prognostically, MRD levels below 0.01% (1 MRD cell per 10,000 bone marrow cells) significantly increase the risk of leukemia relapse, underscoring its clinical relevance [16]. FCM is utilized to differentiate the immunophenotypes typical of leukemic cells from normal cells [55].
PCR is a sensitive method for MRD quantification, detecting gene rearrangements, fusion transcripts, and aberrant genes. Its sensitivity for ALL MRD typically reaches 0.001%. Real-time quantitative PCR (RQ PCR) tracks DNA amplification in real-time using fluorescent probes, while reverse transcription PCR (RT-PCR) processes mRNA to quantify fusion gene transcripts. Bone marrow samples, collected at treatment intervals, are the primary source for PCR-based MRD analysis, though peripheral blood can also be used with sensitivity ≤ 10-5. Despite being costlier and more labor-intensive than FCM, PCR remains the preferred method for MRD detection due to its superior sensitivity [16].
MiRNA profiling techniques
Numerous techniques are utilized for miRNA profiling (Fig. 3). Microarray analysis, including techniques like array comparative genome hybridization (CGH) and single nucleotide polymorphism (SNP) arrays, is used in molecular cytogenetics to detect specific DNA sequences. A microarray contains thousands of DNA probes that can hybridize with matching DNA sequences from a patient’s sample, enabling the analysis of vast regions of the genome. Array CGH compares a patient’s DNA to a reference genome to identify genomic imbalances, such as copy number alterations, through fluorescence-based detection. SNP arrays detect single nucleotide variations, which can be inherited or acquired, with probes identifying both alleles at specific SNP sites. Microarrays can identify genetic variants like copy number changes, aneuploidy, and single base mutations [56].
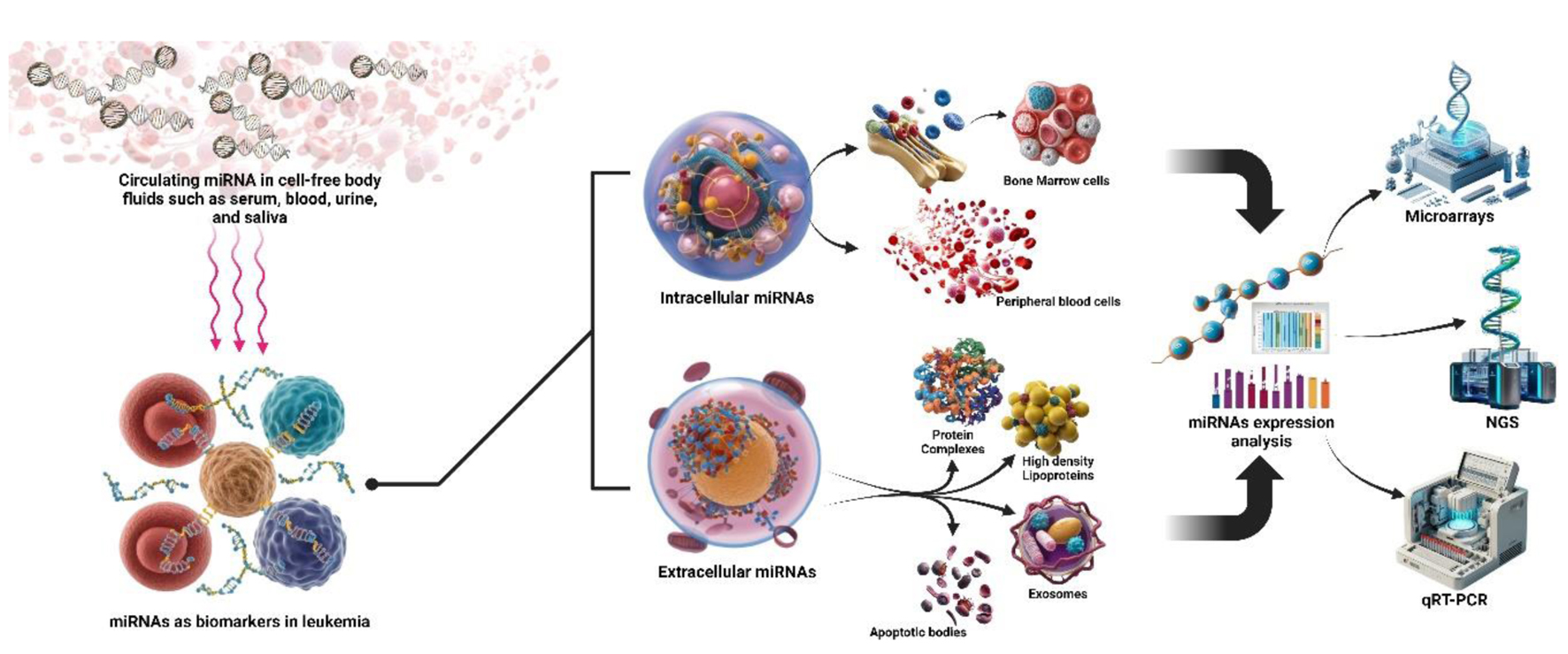 Click for large image | Figure 3. The use of microRNAs (miRNAs) as biomarkers in leukemia, employing both invasive and non-invasive methods for expression analysis. Non-invasive detection involves circulating miRNAs in body fluids like serum, blood, urine, and saliva. Invasive approaches extract intracellular miRNAs from bone marrow and peripheral blood cells. Extracellular miRNAs are also found in protein complexes, high-density lipoproteins, exosomes, and apoptotic bodies. These miRNAs are analyzed using advanced techniques such as microarrays, next generation sequencing (NGS), and quantitative reverse transcription PCR (qRT-PCR), showcasing their potential in leukemia diagnosis and monitoring. |
Moreover, microarray chip analysis, which enables genome-wide gene expression analysis, presents new avenues for identifying markers for MRD studies [55].
NGS is a powerful technique for sequencing DNA fragments, providing precise data on sequences and variations. It is particularly effective in detecting MRD in ALL and AML by identifying epigenetic and clonal alterations with high accuracy. NGS can detect MRD at levels as low as 0.001% to 0.00001%, surpassing the sensitivity of PCR and FCM. The process involves sonication, restriction enzyme application, and PCR amplification, followed by sequencing. NGS is also valuable in diagnosing and monitoring lymphoproliferative disorders by sequencing the lymphoid receptor gene repertoire, aiding in risk stratification and relapse detection [16, 55].
ddPCR is an advanced technique used for MRD detection, offering absolute quantification of target DNA. Unlike RQ-PCR, which provides relative quantification, ddPCR partitions the reaction mixture into approximately 20,000 droplets, each acting as an independent reaction chamber. Fluorescence is measured in each droplet, and the frequency of positive amplifications is analyzed using the Poisson distribution to estimate template concentration [57]. ddPCR has demonstrated high sensitivity in monitoring MRD, particularly in patients post-HSCT. In a study involving 32 JMML patients, ddPCR detected relapses earlier than traditional methods, revealing a critical 1- to 3-month window post-transplantation. Elevated ddPCR-MRD levels within this period significantly predicted relapse and survival outcomes, with a cut-off of 0.465% providing prognostic value. Developed by Vogelstein and Kinzler in 1999 and commercially available since 2011, ddPCR is more sensitive than RQ-PCR, detecting MRD down to one blast cell per 106 cells. However, it is less standardized and requires patient-specific reagents, making it more labor-intensive. Despite these challenges, ddPCR’s ability to provide absolute quantification and its high sensitivity make it a promising alternative for MRD detection, particularly in cases where traditional methods may not be sufficient [58].
| Comparison With Traditional MRD Monitoring Methods | ▴Top |
Monitoring MRD in JMML is crucial for evaluating treatment response and guiding clinical decisions. Traditional methods, such as FCM and PCR-based assays, have been standard for MRD detection. However, recent advancements suggest that miRNA signatures could offer a superior approach. These miRNA signatures are highly sensitive and specific, allowing for the detection of residual leukemia cells even at low levels. Importantly, miRNA signatures can identify JMML-associated miRNAs in patient samples, enabling early detection of disease recurrence [59].
What distinguishes miRNA signatures is their non-invasive nature. Unlike traditional methods that rely on invasive procedures like bone marrow aspirates or biopsies, miRNA signatures can be detected in easily accessible body fluids such as blood or urine. This provides a less invasive, more convenient alternative for patients. Additionally, miRNA signatures offer the potential for precision monitoring, tailored to the unique characteristics of each JMML case, facilitating a personalized approach to treatment [60].
Table 1 [1-3] provides a comparative overview of these traditional MRD monitoring methods and miRNA signatures, highlighting their respective advantages and limitations.
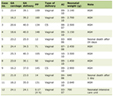 Click to view | Table 1. A Comparative Overview of Traditional MRD Monitoring Methods vs. miRNA Signatures in JMML |
Early MRD detection is critical in preventing relapse, and miRNA signatures have shown the ability to identify MRD at earlier stages than conventional methods [61]. Continued research and clinical validation are essential to establish miRNA signatures as a standard part of routine clinical practice for JMML.
Understanding miRNA-mediated regulatory networks in JMML is crucial for improving diagnosis and treatment strategies. Exosomal miRNAs, which are stable and transportable, have emerged as promising biomarkers for JMML, offering advantages over traditional serum or urine biomarkers due to their higher stability [62]. Additionally, the let-7 miRNA family plays a significant role in JMML pathogenesis, with elevated LIN28B expression, which inhibits let-7, identified in a fetal-like subgroup of JMML. This highlights a key regulatory mechanism that could be targeted for therapeutic interventions. In summary, miRNAs, particularly those in exosomes and the let-7 family, provide valuable insights into JMML pathobiology and offer potential avenues for improved management of the disease [63].
| Impact of MiRNA Dysregulation on Disease Progression | ▴Top |
MiRNA dysregulation plays a critical role in the progression of various diseases by influencing cellular processes such as inflammation and oncogenesis [64]. Altered miRNA expression has been linked to the pathogenesis of numerous diseases, including cancer, where they act as tumor suppressors or oncogenes, impacting tumor development and drug resistance [33]. Dysregulated miRNAs have shown promise as potential biomarkers for early disease diagnosis and monitoring. Furthermore, miRNAs are being explored as therapeutic agents for gene silencing, offering potential for future treatments [65, 66]. Overall, the dysregulation of miRNAs significantly affects disease progression, with ongoing research uncovering new opportunities for diagnosis and therapy.
| Clinical Applications | ▴Top |
Prognostic value of miRNA signatures
The potential of miRNA signatures in cancer prognosis has gained increasing recognition. A signature of 11 miRNAs has been identified as a prognostic marker, aiding clinicians in tailoring treatment plans and predicting cancer outcomes [67]. Additionally, a nine-miRNA immune-related signature has shown effectiveness in predicting prognosis and immune responses, which can influence treatment efficacy and disease progression [68]. Non-invasive blood-based miRNA signatures provide valuable prognostic information, helping predict disease course without the need for invasive procedures [69]. Machine learning techniques have also been applied to miRNA signatures, enhancing the accuracy of prognosis predictions and aiding in personalized treatment strategies [70]. Circulating miRNAs serve as markers for both diagnosis and prognosis, offering insights into disease status and treatment response. A ten-miRNA signature has been shown to be a superior predictor of prognosis, helping clinicians customize treatment plans. While miRNA signatures hold significant promise in cancer research, further research and validation are needed for their clinical application [71, 72].
Monitoring treatment response using miRNA signatures
Monitoring treatment response through miRNA signatures is a growing field, with significant implications in cancer therapy and infectious diseases [73]. MiRNAs have emerged as promising biomarkers for tracking treatment efficacy. Their versatility has been demonstrated in various biological processes, making them appealing candidates for monitoring therapeutic outcomes [74]. Beyond cancer, miRNAs have shown potential in infectious disease treatment monitoring. They are also being explored as therapeutic agents, with ongoing advancements in miRNA therapeutics potentially enhancing their role in treatment response tracking [75, 76]. MiRNAs regulate tumor metabolism, the microenvironment, and immunity, highlighting their multifaceted roles in treatment response. The potential of miRNAs as diagnostic and prognostic tools continues to be investigated, offering hope for improved patient care and treatment management [77, 78].
Personalized treatment approaches based on miRNA profiles
Tailored therapeutic interventions based on miRNA profiles are an emerging field within precision medicine that aims to customize healthcare based on individual patient characteristics [79]. They can identify diseases through unique miRNA profiles and potentially treat conditions by targeting specific miRNAs involved in disease pathways [80]. Accurate miRNA profiling is vital for effective personalized treatment, with tools like “mirTools” aiding in identifying relevant signatures. In cancer, recognizing specific miRNA profiles is especially important, as they can guide personalized treatment strategies, as seen in the case of colon cancer, where specific miRNA profiles may lead to tailored therapies [81]. This technique is expected to be effective in JMML as well.
| Challenges and Future Directions | ▴Top |
The field of miRNA profiling has advanced significantly, yet challenges remain in standardizing techniques. Variations in profiling methods, including microarrays, RNA sequencing, and quantitative PCR, can affect sensitivity, specificity, and data normalization, making cross-study comparisons difficult [82, 83]. To address these challenges, the development of bioinformatics tools for data processing, along with a deeper understanding of miRNA biogenesis, is essential. Overcoming these issues is critical for miRNAs to realize their potential as disease biomarkers and tools for integrating precision medicine data into clinical decision-making, which has a promising development in precision medicine [84]. MiRNAs are implicated in various diseases, including cancer and cardiovascular disorders, and understanding their biogenesis and roles in health and disease is crucial. Bioinformatics tools aid in the analysis of miRNA data, enabling researchers to predict miRNA targets and assess their functional roles [85]. Machine learning is applied to identify disease-associated miRNA signatures, offering new possibilities for personalized treatment, particularly in cancer prognosis [84].
Longitudinal studies are crucial for the dynamic roles of miRNAs, particularly in the context of disease progression and response to treatment in cancers.
Technological advancements, such as high-throughput sequencing and technologies, have significantly improved the sensitivity and specificity of MRD detection. NGS allows comprehensive analysis of miRNA profiles, enabling the detection of low-abundance miRNAs that may indicate MRD in hematological malignancies. However, large-scale clinical trials are needed to validate miRNA-based assays for MRD detection and establish their routine clinical use [86, 87].
Protocols are standardized for miRNA extraction, quantification, and analysis to ensure reproducibility and consistency across different laboratories. This standardization is necessary for the clinical application of miRNA profiling in MRD detection. Combining miRNA data with other omics data, such as genomics and proteomics, enhances the MRD detection. This multi-omics approach provides a more comprehensive view of disease molecular underpinnings and supports better-informed treatment decisions. The integration of ctDNA analysis with miRNA profiling further improves MRD detection by providing complementary insights into tumor biology [88].
In pediatric malignancies like JMML, miRNAs may be used as early prognostic markers, potentially predicting relapse and guiding treatment decisions. Longitudinal monitoring of miRNA levels can aid in early detection of relapse, improving patient outcomes. Exploring miRNA-based therapies, such as miRNA mimics or inhibitors, could offer new avenues for treatment, particularly when combined with existing therapies to enhance efficacy and reduce relapse risk [89, 90].
Overall, the integration of miRNA into MRD detection and its application in hematological malignancies, including JMML, represents a significant advancement in disease management [91, 92]. Continued research, clinical validation, and standardization are essential to fully realize the potential of miRNA-based approaches in precision medicine.
| Ethical Considerations | ▴Top |
In miRNA studies targeting JMML, ensuring patient privacy and informed consent is essential [93]. Ethical guidelines, such as those from the European Leukaemia Net (ELN), emphasize transparency, confidentiality, and participant anonymity. As miRNA research progresses, researchers must adapt their ethical practices to emerging technologies and maintain compliance with data protection laws [94, 95].
Regarding intellectual property rights (IPR) and commercialization, miRNA-based diagnostics and therapies for JMML are gaining importance [96]. While specific discussions on IPR in JMML are limited, related studies on pediatric cancers and RNA therapies highlight the potential for patenting miRNA-based tools [97]. The integration of miRNAs into cancer treatments raises commercialization concerns, including patenting and the transparency of prediction models [98]. Future research will be key in addressing these IPR issues as miRNA technologies continue to evolve.
| Conclusion | ▴Top |
MiRNA signatures hold significant potential as sensitive biomarkers for MRD detection and monitoring in JMML. These small non-coding RNA molecules play critical roles in gene regulation and exhibit both oncogenic and tumor-suppressive functions. In JMML, dysregulated miRNA expression impacts disease progression and clinical outcomes. The identification of specific miRNA signatures correlates with molecular subgroups of JMML, offering promise for personalized, non-invasive MRD monitoring. While current research shows that miRNAs can detect MRD earlier than traditional methods, further validation is required for clinical implementation.
Ethical considerations, such as patient privacy and informed consent, remain critical in miRNA research, while IPR and commercialization will drive advancements in miRNA-based diagnostics and therapies. The integration of miRNA signatures with other technologies, such as single-cell sequencing, could provide deeper insights into JMML biology and improve treatment strategies. Despite promising prospects, miRNA-based MRD monitoring requires standardization and clinical validation to ensure broader adoption.
In conclusion, miRNA signatures offer exciting possibilities for enhancing the clinical management of JMML, improving early detection, treatment monitoring, and patient outcomes. Continued research and validation will be key to realizing their full potential in clinical practice.
Acknowledgments
None to declare.
Financial Disclosure
This research received no external funding.
Conflict of Interest
The authors declare no conflict of interest.
Informed Consent
Not applicable.
Author Contributions
BA wrote, investigated, and reviewed the work. GJ wrote, reviewed, and created figures. RR reviewed the article prior to submission.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
| References | ▴Top |
- Niemeyer CM, Flotho C. Juvenile myelomonocytic leukemia: who's the driver at the wheel? Blood. 2019;133(10):1060-1070.
doi pubmed - Yolanda N, Gunawan S, Mantik MFJ, Veerman AJP. Juvenile myelomonocytic leukemia in a child: a case report of palliative chemotherapy and literature review applied to limited resources centers. Case Rep Hematol. 2022;2022:1185140.
doi pubmed - Sidhu I, Barwe SP, Pillai RK, Gopalakrishnapillai A. Harnessing the power of induced pluripotent stem cells and gene editing technology: therapeutic implications in hematological malignancies. Cells. 2021;10(10):2698.
doi pubmed - Niemeyer CM. JMML genomics and decisions. Hematology Am Soc Hematol Educ Program. 2018;2018(1):307-312.
doi pubmed - Calvo KR, Price S, Braylan RC, Oliveira JB, Lenardo M, Fleisher TA, Rao VK. JMML and RALD (Ras-associated autoimmune leukoproliferative disorder): common genetic etiology yet clinically distinct entities. Blood. 2015;125(18):2753-2758.
doi pubmed - Wintering A, Dvorak CC, Stieglitz E, Loh ML. Juvenile myelomonocytic leukemia in the molecular era: a clinician's guide to diagnosis, risk stratification, and treatment. Blood Adv. 2021;5(22):4783-4793.
doi pubmed - Cortellazzo Wiel L, Pastore S, Taddio A, Tommasini A. A case of uveitis in a patient with juvenile myelomonocytic leukemia successfully treated with adalimumab. J Pediatr Hematol Oncol. 2020;42(5):e373-e376.
doi pubmed - ARUP laboratories resource for education: hematopathology case report: 2-year old male with numerous infections, persistent leukocytosis and monocytosis | University of Utah [Internet]. Available from: https://arup.utah.edu/education/case-reports_hema3/case3_final.php.
- Patnaik MM. How I diagnose and treat chronic myelomonocytic leukemia. Haematologica. 2022;107(7):1503-1517.
doi pubmed - Locatelli F, Niemeyer CM. How I treat juvenile myelomonocytic leukemia. Blood. 2015;125(7):1083-1090.
doi pubmed - Sakashita K, Yoshida N, Muramatsu H, Ohtsuka Y, Watanabe K, Yabe M, Kakuda H, et al. Allogeneic hematopoietic cell transplantation for juvenile myelomonocytic leukemia with a busulfan, fludarabine, and melphalan regimen: JPLSG JMML-11. Transplant Cell Ther. 2024;30(1):105.e1-e10.
doi pubmed - Finana C, Gomez-Molina N, Alonso-Moreno S, Belver L. Genomic and epigenomic landscape of juvenile myelomonocytic leukemia. Cancers (Basel). 2022;14(5):1335.
doi pubmed - Volchkov EV, Khozyainova AA, Gurzhikhanova MK, Larionova IV, Matveev VE, Evseev DA, Ignatova AK, et al. Potential value of high-throughput single-cell DNA sequencing of Juvenile myelomonocytic leukemia: report of two cases. NPJ Syst Biol Appl. 2023;9(1):41.
doi pubmed - Mao S, Lin Y, Qin X, Miao Y, Luo C, Luo C, Wang J, et al. Droplet digital PCR: An effective method for monitoring and prognostic evaluation of minimal residual disease in JMML. Br J Haematol. 2024;204(6):2332-2341.
doi pubmed - Al-Sawaf O, Seymour JF, Kater AP, Fischer K. Should undetectable minimal residual disease be the goal of chronic lymphocytic leukemia therapy? Hematol Oncol Clin North Am. 2021;35(4):775-791.
doi pubmed - Kruse A, Abdel-Azim N, Kim HN, Ruan Y, Phan V, Ogana H, Wang W, et al. Minimal residual disease detection in acute lymphoblastic leukemia. Int J Mol Sci. 2020;21(3):1054.
doi pubmed - Juarez-Avendano G, Mendez-Ramirez N, Luna-Silva NC, Gomez-Almaguer D, Pelayo R, Balandran JC. Molecular and cellular markers for measurable residual disease in acute lymphoblastic leukemia. Bol Med Hosp Infant Mex. 2021;78(3):159-170.
doi pubmed - Sinha R, Dvorak M, Ganesan A, Kalesinskas L, Niemeyer CM, Flotho C, Sakamoto KM, et al. Epigenetic profiling of PTPN11 mutant JMML hematopoietic stem and progenitor cells reveals an aberrant histone landscape. Cancers (Basel). 2023;15(21):5204.
doi pubmed - Leoncini PP, Bertaina A, Papaioannou D, Flotho C, Masetti R, Bresolin S, Menna G, et al. MicroRNA fingerprints in juvenile myelomonocytic leukemia (JMML) identified miR-150-5p as a tumor suppressor and potential target for treatment. Oncotarget. 2016;7(34):55395-55408.
doi pubmed - Vazquez-Urrutia JR, Torres-Bustamante MI, Cerda-Cruz CR, Bravo-Cuellar A, Ortiz-Lazareno PC. The role of miRNA in prostate cancer diagnosis, prognosis and treatment response: a narrative review. Future Oncol. 2023;19(1):77-93.
doi pubmed - Rana S, Valbuena GN, Curry E, Bevan CL, Keun HC. MicroRNAs as biomarkers for prostate cancer prognosis: a systematic review and a systematic reanalysis of public data. Br J Cancer. 2022;126(3):502-513.
doi pubmed - Discovery of novel miRNAs in colorectal cancer: potential biological roles and clinical utility [Internet]. Available from: https://www.mdpi.com/2311-553X/9/6/65?utm_source=chatgpt.com.
- Long non-coding RNAs as novel therapeutic targets in juvenile myelomonocytic leukemia. Scientific Reports [Internet]. Available from: https://www.nature.com/articles/s41598-021-82509-5.
- Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130(11):1290-1301.
doi pubmed - Fletcher D, Brown E, Javadala J, Uysal-Onganer P, Guinn BA. microRNA expression in acute myeloid leukaemia: New targets for therapy? EJHaem. 2022;3(3):596-608.
doi pubmed - Liao Q, Wang B, Li X, Jiang G. miRNAs in acute myeloid leukemia. Oncotarget. 2017;8(2):3666-3682.
doi pubmed - Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117(4):1121-1129.
doi pubmed - Garofalo M, Croce CM. microRNAs: Master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25-43.
doi pubmed - Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102-114.
doi pubmed - The James - OSUCCC [Internet]. [cited 2023 Aug 23]. microRNA - What it is and how it works | OSUCCC - James. Available from: https://cancer.osu.edu/microrna.
- Ranganathan K, Sivasankar V. MicroRNAs - biology and clinical applications. J Oral Maxillofac Pathol. 2014;18(2):229-234.
doi pubmed - Lam JKW, Chow MYT, Zhang Y, Leung SWS. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther - Nucleic Acids. 2015;4:e252.
doi - Nies YH, Mohamad Najib NH, Lim WL, Kamaruzzaman MA, Yahaya MF, Teoh SL. MicroRNA dysregulation in Parkinson's disease: a narrative review. Front Neurosci. 2021;15:660379.
doi pubmed - Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004.
doi pubmed - Ardekani AM, Naeini MM. The role of MicroRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161-179.
pubmed - Peng Y, Zhang X, Feng X, Fan X, Jin Z. The crosstalk between microRNAs and the Wnt/beta-catenin signaling pathway in cancer. Oncotarget. 2017;8(8):14089-14106.
doi pubmed - Singh S, Shyamal S, Das A, Panda AC. Global identification of mRNA-interacting circular RNAs by CLiPPR-Seq. Nucleic Acids Res. 2024;52(6):e29.
doi pubmed - Wang XF, Yu CQ, Li LP, You ZH, Huang WZ, Li YC, Ren ZH, et al. KGDCMI: a new approach for predicting circRNA-miRNA interactions from multi-source information extraction and deep learning. Front Genet. 2022;13:958096.
doi pubmed - Sakshi S, Jayasuriya R, Ganesan K, Xu B, Ramkumar KM. Role of circRNA-miRNA-mRNA interaction network in diabetes and its associated complications. Mol Ther Nucleic Acids. 2021;26:1291-1302.
doi pubmed - Bizzarri AR, Cannistraro S. Direct interaction of miRNA and circRNA with the Oncosuppressor p53: an intriguing perspective in cancer research. Cancers (Basel). 2021;13(23):6108.
doi pubmed - Lei Y, Chen L, Zhang G, Shan A, Ye C, Liang B, Sun J, et al. MicroRNAs target the Wnt/beta-catenin signaling pathway to regulate epithelial-mesenchymal transition in cancer (Review). Oncol Rep. 2020;44(4):1299-1313.
doi pubmed - Sahu AK, Said MS, Hingamire T, Gaur M, Khan A, Shanmugam D, Barvkar VT, et al. Approach to nigericin derivatives and their therapeutic potential. RSC Adv. 2020;10(70):43085-43091.
doi pubmed - Han Z, Rosen ST, Querfeld C. Targeting microRNA in hematologic malignancies. Curr Opin Oncol. 2020;32(5):535-544.
doi pubmed - Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of microRNA expression in cancer. Int J Mol Sci. 2020;21(5):1723.
doi pubmed - Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287-314.
doi pubmed - Mulero-Navarro S, Sevilla A, Roman AC, Lee DF, D'Souza SL, Pardo S, Riess I, et al. Myeloid dysregulation in a human induced pluripotent stem cell model of PTPN11-associated juvenile myelomonocytic leukemia. Cell Rep. 2015;13(3):504-515.
doi pubmed - Helsmoortel HH, Bresolin S, Lammens T, Cave H, Noellke P, Caye A, Ghazavi F, et al. LIN28B overexpression defines a novel fetal-like subgroup of juvenile myelomonocytic leukemia. Blood. 2016;127(9):1163-1172.
doi pubmed - Szczepanek J. Role of microRNA dysregulation in childhood acute leukemias: Diagnostics, monitoring and therapeutics: A comprehensive review. World J Clin Oncol. 2020;11(6):348-369.
doi pubmed - Mendiola-Soto DK, Barcenas-Lopez DA, Perez-Amado CJ, Cruz-Miranda GM, Mejia-Arangure JM, Ramirez-Bello J, Hidalgo-Miranda A, et al. MiRNAs in hematopoiesis and acute lymphoblastic leukemia. Int J Mol Sci. 2023;24(6):5436.
doi pubmed - Lv M, Zhu S, Peng H, Cheng Z, Zhang G, Wang Z. B-cell acute lymphoblastic leukemia-related microRNAs: uncovering their diverse and special roles. Am J Cancer Res. 2021;11(4):1104-1120.
pubmed - Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S. The therapeutic potential of microRNAs in cancer. Cancer J. 2012;18(3):275-284.
doi pubmed - de Oliveira JC, Brassesco MS, Scrideli CA, Tone LG, Narendran A. MicroRNA expression and activity in pediatric acute lymphoblastic leukemia (ALL). Pediatr Blood Cancer. 2012;59(4):599-604.
doi pubmed - Xing Y, Wang Z, Lu Z, Xia J, Xie Z, Jiao M, Liu R, et al. MicroRNAs: immune modulators in cancer immunotherapy. Immunother Adv. 2021;1(1):ltab006.
doi pubmed - Yuan Y, Zhang H, Liu X, Lu Z, Li G, Lu M, Tao X. MicroRNA signatures predict prognosis of patients with glioblastoma multiforme through the Cancer Genome Atlas. Oncotarget. 2017;8(35):58386-58393.
doi pubmed - Alyamani NM. Minimal residual disease in acute leukemia based on the insight of molecular genetics monitoring. Int J Adv Appl Sci. 2023;10(5):72-85.
- Clinical-Genome-Analysis-Evidence-Review-2020-FINAL.pdf [Internet]. Available from: https://www.sglc.scot.nhs.uk/wp-content/uploads/2020/08/Clinical-Genome-Analysis-Evidence-Review-2020-FINAL.pdf.
- British Journal of Haematology | Wiley Online Library [Internet]. Available from: https://onlinelibrary.wiley.com/doi/epdf/10.1111/bjh.19465.
- Della Starza I, De Novi LA, Elia L, Bellomarino V, Beldinanzi M, Soscia R, Cardinali D, et al. Optimizing molecular minimal residual disease analysis in adult acute lymphoblastic leukemia. Cancers (Basel). 2023;15(2):374.
doi pubmed - Ommen HB. Monitoring minimal residual disease in acute myeloid leukaemia: a review of the current evolving strategies. Ther Adv Hematol. 2016;7(1):3-16.
doi pubmed - Minimal residual disease testing | Choose the right test [Internet]. Available from: https://arupconsult.com/content/minimal-residual-disease-testing.
- 2017 Aspho Abstracts. Pediatr Blood Cancer. 2017;64(S1):e26591.
- Galardi A, Colletti M, Di Paolo V, Vitullo P, Antonetti L, Russo I, Di Giannatale A. Exosomal MiRNAs in Pediatric Cancers. Int J Mol Sci. 2019;20(18):4600
doi - Yazarlou F, Kadkhoda S, Ghafouri-Fard S. Emerging role of let-7 family in the pathogenesis of hematological malignancies. Biomed Pharmacother. 2021;144:112334.
doi pubmed - Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18(3):215-222.
doi pubmed - Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. 2019;11(1):25.
doi pubmed - Visacri MB, Nicoletti AS, Pincinato EC, Loren P, Saavedra N, Saavedra K, Salazar LA, et al. Role of miRNAs as biomarkers of COVID-19: a scoping review of the status and future directions for research in this field. Biomark Med. 2021;15(18):1785-1795.
doi pubmed - Lu J, Liang J, Xu M, Wu Z, Cheng W, Wu J. Identification of an eleven-miRNA signature to predict the prognosis of endometrial cancer. Bioengineered. 2021;12(1):4201-4216.
doi pubmed - Chen S, Gao C, Wu Y, Huang Z. Identification of prognostic miRNA signature and lymph node metastasis-related key genes in cervical cancer. Front Pharmacol. 2020;11:544.
doi pubmed - Rajakumar T, Horos R, Jehn J, Schenz J, Muley T, Pelea O, Hofmann S, et al. A blood-based miRNA signature with prognostic value for overall survival in advanced stage non-small cell lung cancer treated with immunotherapy. NPJ Precis Oncol. 2022;6(1):19.
doi pubmed - Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8-17.
doi pubmed - Terrinoni A, Calabrese C, Basso D, Aita A, Caporali S, Plebani M, Bernardini S. The circulating miRNAs as diagnostic and prognostic markers. Clin Chem Lab Med. 2019;57(7):932-953.
doi pubmed - Ma R, Zhao Y, He M, Zhao H, Zhang Y, Zhou S, Gao M, et al. Identifying a ten-microRNA signature as a superior prognosis biomarker in colon adenocarcinoma. Cancer Cell Int. 2019;19:360.
doi pubmed - Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016;231(1):25-30.
doi pubmed - Tribolet L, Kerr E, Cowled C, Bean AGD, Stewart CR, Dearnley M, Farr RJ. MicroRNA biomarkers for infectious diseases: from basic research to biosensing. Front Microbiol. 2020;11:1197.
doi pubmed - Reda El Sayed S, Cristante J, Guyon L, Denis J, Chabre O, Cherradi N. MicroRNA therapeutics in cancer: current advances and challenges. Cancers (Basel). 2021;13(11).
doi pubmed - Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics. 2018;10:59.
doi pubmed - The role of MicroRNA in hepatocellular carcinoma | JHC [Internet]. Available from: https://www.dovepress.com/micrornas-as-modulators-of-tumor-metabolism-microenvironment-and-immun-peer-reviewed-fulltext-article-JHC.
- Cancer Drug Resistance [Internet]. Available from: https://cdrjournal.com/article/view/3354.
- Sempere LF, Azmi AS, Moore A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip Rev RNA. 2021;12(6):e1662.
doi pubmed - Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun Z, et al. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38(Web Server issue):W392-397.
doi pubmed - Poniewierska-Baran A, Zadroga L, Danilyan E, Malkowska P, Niedzwiedzka-Rystwej P, Pawlik A. MicroRNA as a diagnostic tool, therapeutic target and potential biomarker in cutaneous malignant melanoma detection-narrative review. Int J Mol Sci. 2023;24(6):5386.
doi pubmed - Cirera S, Andersen-Ranberg EU, Langkilde S, Aaquist M, Gredal H. Challenges and standardization of microRNA profiling in serum and cerebrospinal fluid in dogs suffering from non-infectious inflammatory CNS disease. Acta Vet Scand. 2019;61(1):57.
doi pubmed - Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G. Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 2019;20(5):1836-1852.
doi pubmed - O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402.
doi pubmed - Saini V, Dawar R, Suneja S, Gangopadhyay S, Kaur C. Can microRNA become next-generation tools in molecular diagnostics and therapeutics? A systematic review. Egypt J Med Hum Genet. 2021;22(1):4.
- Luna Buitrago D, Lovering RC, Caporali A. Insights into online microRNA bioinformatics tools. Noncoding RNA. 2023;9(2):18.
doi pubmed - Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13.
doi pubmed - Abu-Halima M, Keller A, Becker LS, Fischer U, Engel A, Ludwig N, Kern F, et al. Dynamic and static circulating cancer microRNA biomarkers - a validation study. RNA Biol. 2023;20(1):1-9.
doi pubmed - Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12(1):8.
doi pubmed - Li W, Wang F, Guo R, Bian Z, Song Y. Targeting macrophages in hematological malignancies: recent advances and future directions. J Hematol Oncol. 2022;15(1):110.
doi pubmed - Tang L, Huang Z, Mei H, Hu Y. Immunotherapy in hematologic malignancies: achievements, challenges and future prospects. Signal Transduct Target Ther. 2023;8(1):306.
doi pubmed - New directions in the diagnosis and treatment of hematologic cancers: highlights from international medical meetings - Physician Resources | Fox Chase Cancer Center [Internet]. Available from: https://physicianresources.foxchase.org/symposium/new-directions-in-the-diagnosis-and-treatment-of- -cancers-highlights-from-international-medical-meetings/flatDisplayPageSymposium.
- Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, Dombret H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447.
doi pubmed - Mohrmann L, Werner M, Oles M, Mock A, Uhrig S, Jahn A, Kreutzfeldt S, et al. Comprehensive genomic and epigenomic analysis in cancer of unknown primary guides molecularly-informed therapies despite heterogeneity. Nat Commun. 2022;13(1):4485.
doi pubmed - Galardi A, Colletti M, Di Paolo V, Vitullo P, Antonetti L, Russo I, Di Giannatale A. Exosomal MiRNAs in pediatric cancers. Int J Mol Sci. 2019;20(18):4600.
doi pubmed - Jafari M, Kadkhodazadeh M, Shapourabadi MB, Goradel NH, Shokrgozar MA, Arashkia A, Abdoli S, et al. Immunovirotherapy: the role of antibody based therapeutics combination with oncolytic viruses. Front Immunol. 2022;13:1012806.
doi pubmed - Zeiser R, Beelen DW, Bethge W, Bornhauser M, Bug G, Burchert A, Christopeit M, et al. Biology-driven approaches to prevent and treat relapse of myeloid neoplasia after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2019;25(4):e128-e140.
doi pubmed - Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, Vickers AJ, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1-73.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.