| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website https://jh.elmerpub.com |
Review
Volume 14, Number 4, August 2025, pages 165-173
Beyond the Blood Cell: The Emerging Role of Cell-Free DNA in Transfusion Medicine
Jackson M. Wahmana, Rhoda X. Hijazia, Elizabeth Duncana, Petra Rocica, Dominica Moussokia, Hosam G. Abdelhadya, b
aDepartment of Physiology and Pharmacology, College of Osteopathic Medicine, Sam Houston State University, Conroe, TX, USA
bCorresponding Author: Hosam G. Abdelhady, Department of Physiology and Pharmacology, College of Osteopathic Medicine, Sam Houston State University, Conroe, TX 77304, USA
Manuscript submitted March 24, 2024, accepted May 29, 2025, published online July 8, 2025
Short title: Cell-Free DNA in Transfusion Medicine
doi: https://doi.org/10.14740/jh2064
- Abstract
- Introduction
- Methodology
- cfDNA Transfer and Microchimerism
- Immunologic and Pathological Associations of cfDNA
- Dietary and Environmental DNA: Implications for Transfusion Medicine
- Conclusions
- References
| Abstract | ▴Top |
Cell-free DNA (cfDNA) consists of fragmented nuclear and mitochondrial DNA circulating in the bloodstream, primarily originating from hematopoietic cells. While cfDNA analysis has transformed diagnostic medicine, its presence in transfused blood products introduces emerging clinical concerns. Donor-derived cfDNA may persist in transfusion recipients and contribute to transfusion-associated microchimerism, defined as the long-term presence of donor genetic material in recipient tissues or circulation. These fragments have potential to integrate into the host genome, modify DNA methylation and histone structure, and activate innate immune pathways such as Toll-like receptors. In addition to nuclear and mitochondrial sources, cfDNA in transfused blood may include environmental or dietary DNA acquired by donors, further influencing immune regulation. Current leukoreduction methods do not eliminate cfDNA or prevent microchimerism. This review synthesizes current evidence regarding the persistence, genomic integration, and immunologic impact of cfDNA in transfusion recipients. The findings highlight an urgent need for further investigation and refinement of blood processing practices to ensure transfusion safety and protect recipient health.
Keywords: Cell-free DNA; Microchimerism; Allogeneic transfusion; PRBCs; Epigenetics; Gene transfer; Immune activation
| Introduction | ▴Top |
Cell-free DNA (cfDNA): origins and biological properties
cfDNA consists of small and large fragments of double-stranded DNA or chromatin, typically ranging from 50 to 200 base pairs long, released from cells throughout the body [1, 2]. These fragments can originate from both mitochondrial and nuclear DNA and may be derived from either healthy or diseased cells [3]. Recent studies have demonstrated that cfDNA in the bloodstream predominantly exists in nucleosome-associated form, reflecting the physiological processes of chromatin degradation (breakdown of DNA-protein complexes during cell death) and nucleosome stabilization (DNA wrapped around histone proteins, protecting it from enzymatic degradation) [4-8].
While initially discovered in blood plasma, cfDNA has been found in various bodily fluids. In healthy individuals, the majority of cfDNA comes from the hematopoietic system, with around 55% derived from white blood cells (WBCs), 30% from erythrocyte progenitors, and 10% from vascular endothelial cells [1, 9, 10]. Non-hematopoietic sources, including hepatocytes, heart, and lung tissues, contribute smaller but detectable amounts (1-5%) [11]. The body utilizes mechanisms such as phagocytosis of apoptotic and necrotic cells to remove cellular debris and limit cfDNA release, though clearance mechanisms remain incompletely understood [12]. Beyond these passive pathways, viable cells actively release cfDNA through mechanisms including neutrophil extracellular traps (NETs) [13, 14]. When the rate of cfDNA release exceeds the capacity of enzymatic degradation, cfDNA may persist in circulation and accumulate systemically [15, 16]. Histone-bound cfDNA further resists enzymatic clearance, as nucleosome packaging enhances its structural integrity and shields it from DNase activity [16, 17].
NETs are expansive, DNA-protein lattices extruded by activated neutrophils to immobilize and neutralize invading pathogens, including bacteria, fungi, and viruses [13, 14, 18]. The DNA released during NETosis contributes significantly to the circulating cfDNA pool and exerts immunomodulatory effects, particularly in the setting of organ transplantation and immune rejection [19, 20]. For example, in liver transplantation, NET-derived cfDNA correlates with ischemia-reperfusion injury and coagulation activation [13]. Mitochondrial cfDNA (cfDNA-mt) contains cytosine-phosphate-guanine (CpG) sites that exhibit inter-individual methylation variability, with hypomethylated CpG-rich regions linked to transcriptional activation [21-23]. Trauma patients and individuals with critical illness exhibit elevated levels of CpG-rich cfDNA-mt, which have been correlated with systemic inflammation and adverse clinical outcomes [13, 24].
cfDNA serves as a biomarker of chromosomal instability and tissue damage, reflecting pre-existing genomic alterations such as double-strand breaks or chromosomal rearrangements in parent cells (e.g., tumors or apoptotic leukocytes) [25-27]. Integration into recipient genomes occurs at fragile sites via non-homologous end joining (NHEJ), a process observed in cancer and transfusion-associated microchimerism (TA-MC) [11, 28]. Transfused cfDNA may persist as microchimerism, activate Toll-like receptor 9 (TLR9)-mediated inflammation via CpG motifs [13, 22], or modulate immune responses [28]. These potential effects of cfDNA on the host including genomic integration, immune activation, epigenetic modification, and microchimerism are summarized in Figure 1.
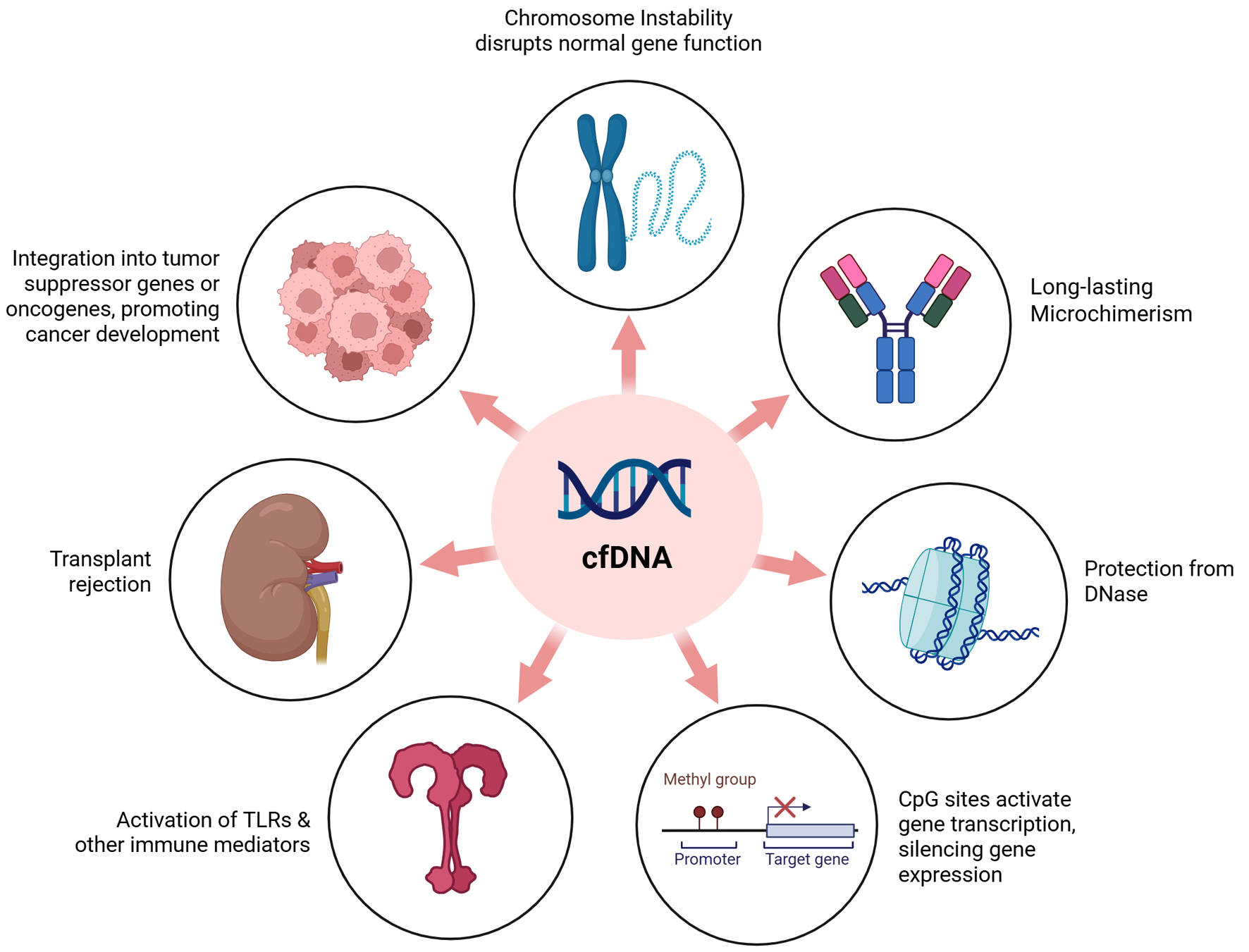 Click for large image | Figure 1. Theorized effects of cfDNA on the transfusion recipient. The impact of transfused cfDNA on the recipient may vary widely depending on factors such as cfDNA concentration, the recipient’s genomic background and lifestyle, and the cellular origin of the cfDNA. It is hypothesized that cfDNA bound to histones may evade DNase degradation and potentially integrate into the host genome. This integration could influence gene regulation by promoting transcriptional activation or silencing. cfDNA has also been associated with genomic instability in theoretical models. If not effectively cleared by the immune system, donor cfDNA may contribute to enhanced immune activation, potentially triggering responses linked to transplant rejection or the activation of pathways involving tumor suppressor genes, oncogenes, or Toll-like receptors. cfDNA: cell-free DNA; CpG: cytosine-phosphate-guanine; TLR: Toll-like receptor. |
The presence of cfDNA in blood products: sources, implications, and concerns
Packed red blood cells (PRBCs) are among the most frequently used blood transfusion products. These preparations primarily contain red blood cells (RBCs) and a relatively low number of donor-derived WBCs. Leukoreduction, a filtration process designed to remove WBCs before transfusion, is widely implemented to reduce the risk of immunologic complications [29]. While leukoreduction techniques can help limit the WBC count in PRBC products, they are not always universally applied, and acceptable WBC levels in blood products vary based on different standards [30]. Notably, studies have shown that leukoreduction has not been shown to affect the presence or duration of TA-MC [30].
A key concern in transfusion medicine is the presence of cfDNA in blood products. While the primary source of cfDNA is donor WBCs, additional contributions may come from apoptotic cells derived from various tissues, NETs, stem cells, and even dietary or environmental DNA [1, 9, 10, 16, 31-34]. While mature RBCs lack nuclei and mitochondria, and thus do not contain DNA, they are rich in various RNA species [35]. However, studies have shown that the predominant nucleic acids in blood products are double-stranded DNA rather than RNA [16, 30, 31]. These potential contributors to the circulating cfDNA pool are summarized in Figure 2.
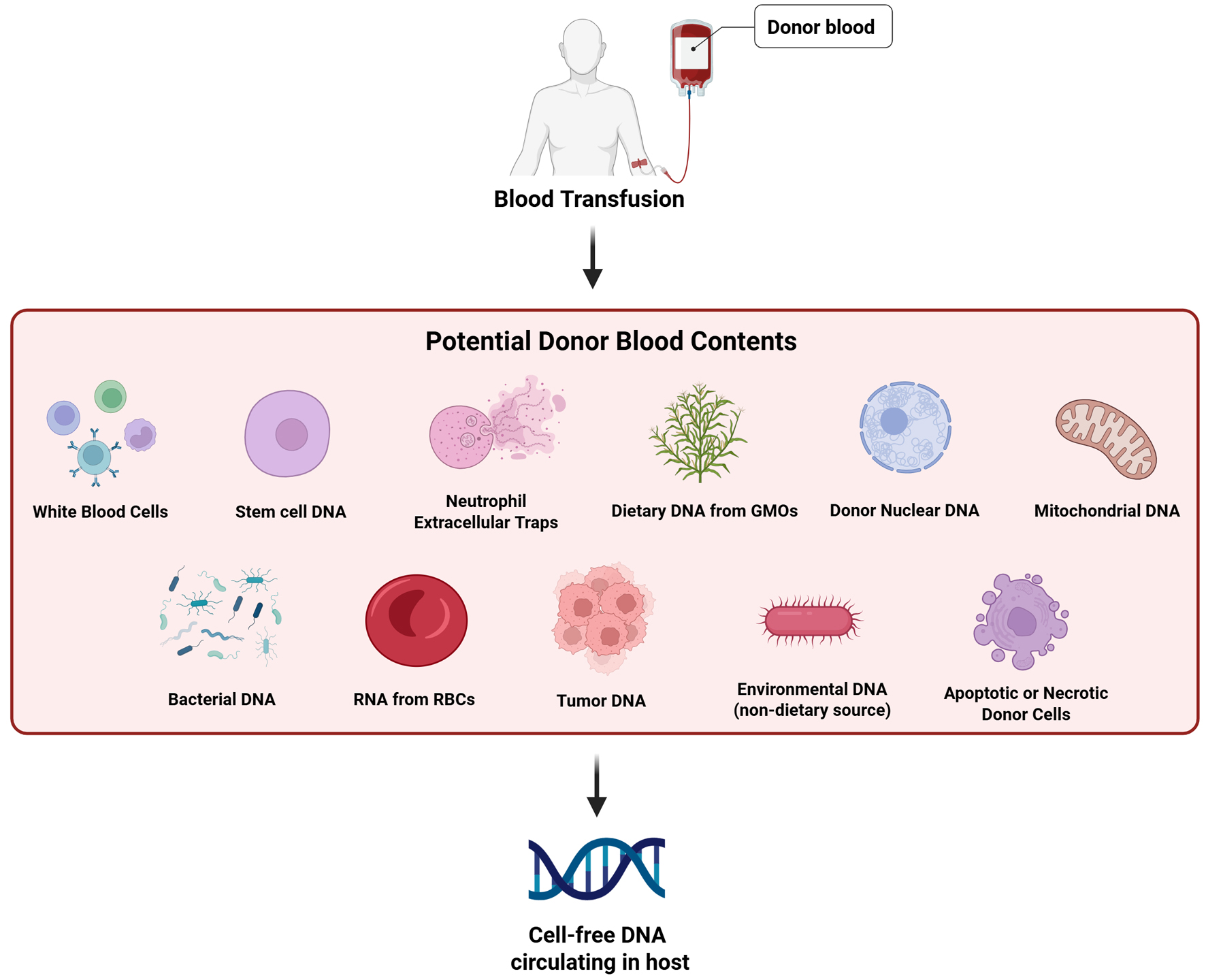 Click for large image | Figure 2. Potential sources of donor-derived cfDNA in transfused blood. Donor blood may contain various sources of cfDNA, including DNA from white blood cells, stem cells, NETs, dietary DNA (e.g., from GMOs), nuclear DNA, and mt-cfDNA. These fragments can persist in the recipient’s circulation post-transfusion, despite leukoreduction, and may contribute to downstream immunologic or epigenetic effects. cfDNA: cell-free DNA; cfDNA-mt: mitochondrial cell-free DNA; GMOs: genetically modified organisms; NETs: neutrophil extracellular traps. |
Beyond cellular origin, several logistical and technical variables influence the presence of cfDNA in transfused blood. Although PRBCs are stored for variable durations to meet clinical demand, studies show that neither storage time nor processing significantly affects cfDNA levels [36, 37]. Consequently, modifications to storage and processing techniques are unlikely to prevent cfDNA transmission during transfusion. While cfDNA detection has advanced noninvasive prenatal testing, transplant monitoring, and cancer diagnostics [38-42], its inadvertent transfer through transfusion remains a matter of concern.
| Methodology | ▴Top |
This narrative review synthesized current and foundational literature on cfDNA in transfusion medicine. A comprehensive search of PubMed, Scopus, Web of Science, and Google Scholar was conducted using combinations of the following terms: “cell-free DNA,” “cfDNA,” “circulating free DNA,” “blood transfusion,” “packed red blood cells,” “PRBCs,” “microchimerism,” “donor-derived DNA,” “horizontal gene transfer,” “epigenetics,” and “cfDNA integration.” Studies were selected through title and abstract screening, followed by full-text review. Research from the last 10 years was prioritized, with earlier seminal works included for context. Only peer-reviewed in vitro, in vivo, and observational studies were considered; conference abstracts and preprints were excluded. No formal analysis was performed due to the narrative nature of this review. Where findings conflicted, differing perspectives were included with attention to methodological quality. The evidence quality was assessed by considering study design, sample size, methodological rigor, and consistency with other published findings.
| cfDNA Transfer and Microchimerism | ▴Top |
Microchimerism is defined as the long-term persistence of genetically distinct cells or cfDNA within an individual, originating from another organism. This phenomenon occurs bidirectionally through natural processes (e.g., pregnancy, twinning) or artificial routes (e.g., blood transfusion, organ transplantation) [42-45]. In transfusion medicine, TA-MC refers specifically to donor-derived genetic material (cells or cfDNA) that persists in recipients, even when leukoreduced blood products are used [45-47].
The integration of cfDNA and its impact on gene expression and genomic stability are considered crucial factors contributing to these long-term effects [43, 48]. For example, when Khaki Campbell duck DNA was injected into Pekin ducks, unexpected modifications occurred in nine out of 12 treated ducks, while no changes were observed in 24 control ducks [49]. These modified traits were passed down to descendants for up to three generations, suggesting that the DNA-induced changes were both stable and transmissible [49]. Although conducted decades ago, this experiment offers early proof of principle for horizontal DNA uptake leading to stable, transmissible genetic changes.
Building on these foundational insights, recent research has highlighted the clinical relevance of cfDNA in modern medicine. In transplantation, donor-derived cfDNA (dd-cfDNA) is now recognized as a sensitive biomarker for early detection of allograft injury and rejection, outperforming traditional markers such as serum creatinine. For example, dd-cfDNA levels > 1% demonstrate high diagnostic accuracy for acute rejection in pediatric renal transplants [50], while elevated dd-cfDNA in kidney allograft recipients strongly correlates with antibody-mediated and T-cell-mediated rejection, independent of standard clinical parameters [51]. In transfusion medicine, in vitro studies demonstrate that transfused cfDNA may alter recipient cell behavior and may persist and expand for months or longer, particularly when genomic integration or microchimerism occurs [43, 46, 52]. Despite these advances, the long-term clinical consequences of dd-cfDNA in transfusion recipients remain poorly understood [53].
| Immunologic and Pathological Associations of cfDNA | ▴Top |
Emerging evidence shows that cfDNA can integrate into host genomes and alter gene expression, contributing to durable microchimerism [43, 54]. In vitro studies have demonstrated that the internalized cfDNA may influence gene regulation, particularly by inducing pro-inflammatory genes in the cells that have absorbed the cfDNA [54-59]. Though best known in bacteria for facilitating the spread of antibiotic resistance, in human cells, horizontal gene transfer (HGT) has emerged as a relevant model for understanding cfDNA uptake and functional integration [60, 61].
Adding to this growing body of evidence, donor DNA has been detected not only in recipient blood but also in epithelial cells (e.g., buccal mucosa) unrelated to transfusion [62]. Early theories attributed this to stem cell plasticity, but contemporary evidence implicates gene transfer (GT) as the primary mechanism, enabling even non-hematopoietic cells to acquire donor DNA [63]. Seminal work by Garcia-Olmo et al demonstrated that plasma from colorectal cancer patients could transfer human oncogenes such as K-ras and p53 to murine fibroblasts, which subsequently induced tumor formation in immunodeficient mice [63]. This evidence underpins the genometastasis theory, which proposes that circulating tumor DNA has the potential to induce malignant transformation in distant, susceptible cells [63, 64]. More recently, Salazar et al showed that hepatic progenitor cells co-cultured with colorectal cancer cells could acquire the KrasG12D mutation and exhibit tumorigenic behavior, reinforcing the concept that horizontally transferred cfDNA can have lasting functional effects in recipient tissues [64].
Beyond oncogenic transformation, the immune consequences of cfDNA exposure are also significant. Once internalized, dd-cfDNA may engage innate immune sensors such as TLR-9, triggering nuclear factor-kappaB (NF-κB) activation and downstream expression of pro-inflammatory cytokines including interleukin (IL)-1β and C-X-C chemokine ligand 8 (CXCL8) [65, 66]. These immune responses mirror those observed in infectious and neoplastic conditions, further underscoring the immunogenic potential of transfused cfDNA (Fig. 1).
It is further reported that a small portion of cfDNA can circulate as nucleosomes, which consist of DNA wound tightly around histones, that may exert biological effects, including modulation of immune responses and regulation of gene expression [54, 67-69]. Rather than being a passive biomarker, cfDNA plays a dynamic role in maintaining immune balance. For example, cfDNA clearance in plasma is indispensable for maintaining immune homeostasis, and its accumulation can trigger pro-inflammatory pathways and complement activation [68]. In vitro studies show that NETs release high-molecular-weight DNA (1 - 30 kbp) that serum nucleases rapidly degrade into mononucleosomal fragments (81.3% of remaining DNA after 24 h), mirroring the size profile of cfDNA in inflammatory diseases [54].
The broader pathological relevance of cfDNA has been demonstrated across diverse clinical contexts. Elevated cfDNA levels have been observed in malignancy, autoimmune diseases, trauma, sepsis, psychiatric and neurodegenerative conditions, and organ dysfunction [70-76]. For instance, Lopes et al reported that cfDNA concentrations rise substantially during and after major surgery, particularly in patients with advanced disease, with shorter DNA fragments predominating [77]. In bacterial sepsis, increased cfDNA is strongly linked to poor prognosis, likely due to TLR activation and subsequent cytokine storm [54, 58]. This is likely due to activation of TLRs and the resulting cytokine storm, a cascade linked to splenic and acute lung injury and prolongation of neutrophil lifespan [60]. As a recognized damage-associated molecular pattern (DAMP), cfDNA can amplify or mitigate disease progression through its interactions with pattern recognition receptors [77, 78].
Although these findings stem from other pathological conditions and non-transfusion settings, they raise important concerns: if endogenous cfDNA can contribute to such profound pathophysiology, the introduction of dd-cfDNA via transfusion may carry underappreciated risks, particularly in susceptible recipients. As transfusion remains a cornerstone of supportive care, these parallels underscore the urgency of evaluating the biological activity and clinical impact of cfDNA in transfusion medicine.
| Dietary and Environmental DNA: Implications for Transfusion Medicine | ▴Top |
While much of the focus has been on cfDNA derived from human cellular sources, emerging evidence suggests that environmental and dietary DNA may also play a role in transfusion-related exposures [79, 80]. This broader category of cfDNA introduces new dimensions to consider, particularly when evaluating the origins, persistence, and biological activity of genetic material in transfused blood. The gastrointestinal tract serves as the primary route of natural exposure to foreign DNA through food consumption. Research shows that 1-2% of ingested DNA can survive digestion, penetrate the intestinal wall, and reach the nuclei of leukocytes, spleen, and liver cells and may occasionally be integrated into the host genome [80, 81]. Animal studies have shown that dietary DNA can also traverse the placental barrier and persist in fetal tissues for extended periods, prompting concerns about the long-term effects of environmental DNA exposure [80]. In humans, dietary cfDNA, stemming from donor dietary habits, may resist degradation, cross the intestinal barrier, enter the bloodstream and be present in transfused blood products [80, 82]. A large study of maternal blood samples found plant-derived DNA in circulation, with higher levels observed in patients experiencing systemic inflammation, suggesting that dietary cfDNA may play a role in modulating immune responses [82].
Concerns about genetically modified organisms (GMOs) in the diet have centered on the potential for HGT of antibiotic resistance genes or other transgenic elements to human cells or gut microbiota [83]. While heat treatment of foods might reduce the risk of DNA transfer, it also causes significant DNA base damage, which may increase genotoxicity and DNA repair activity in recipient tissues [77, 80]. Unlike food, blood products cannot be heat treated without destroying their viability, so any cfDNA present in donor blood, including that derived from GMOs, may be transfused directly to recipients [20] (Fig. 2).
Once introduced to the recipient, donor cfDNA may integrate into host genomes via homologous recombination (HR) or NHEJ, both of which may disrupt gene function or genomic stability [43, 84, 85]. Integration of cfDNA has been shown to alter DNA methylation and transcription patterns, potentially driving oncogenic or other epigenetic changes [86]. For example, cfDNA from leukemic cells can induce DNA damage and apoptosis in stromal cells [37, 87]. Moreover, environmental factors such as diet and psychological stress may further influence DNA methylation patterns in recipients, compounding the potential for epigenetic dysregulation [88, 89].
Together, these findings support the plausibility that both dietary and transfusion-derived cfDNA could influence recipient health. This possibility warrants further exploration into the biological fate and downstream consequences of cfDNA exposure in transfusion settings, especially as clinical use of blood products grows more widespread.
| Conclusions | ▴Top |
The presence of cfDNA in blood products poses significant concerns for transfusion medicine. cfDNA, which includes both mitochondrial and nuclear DNA fragments, originates from leukocytes and possibly donor stem cells in transfused blood. Despite leukoreduction techniques aimed at reducing WBC content, cfDNA levels can persist for extended periods post-transfusion, ranging from minutes to years, contributing to TA-MC. This integration into the recipient’s genome can lead to genomic instability, increased mutation susceptibility, and disruption of gene function. Furthermore, cfDNA alters the epigenome by modifying DNA methylation patterns and histone configurations, potentially initiating oncogenic pathways. It also modulates inflammatory and immune responses, stimulating pro-inflammatory gene expression. cfDNA can resist degradation, allowing it to be internalized by cells, translocate to the nucleus, and affect gene regulation. This persistence raises the risk of long-term immune modulation and latent inflammatory responses. Additionally, cfDNA fragments from tumor cells could accelerate tumorigenesis. Current processing methods have limited impact on cfDNA persistence, highlighting the need for improved management strategies. Future research should focus on the mechanisms of cfDNA integration, its epigenetic effects, and the long-term outcomes of transfusion-related cfDNA transfer.
Acknowledgments
None to declare.
Financial Disclosure
Article Publishing Charge (APC) was funded by College of Osteopathic Medicine, Sam Houston State University.
Conflict of Interest
The authors declare that they have no competing interests.
Author Contributions
Jackson M. Wahman led the narrative review, performed the majority of the literature search and synthesis, and drafted the initial manuscript. Rhoda Hijazi contributed to manuscript development, analysis of sources, and critical revisions. Elizabeth Duncan designed and prepared the figures. Petra Rocic provided expert review and editorial feedback. Dominica Moussoki supported the literature search and source curation. Hosam G. Abdelhady conceptualized the project, served as the corresponding author, and oversaw all phases of development and final approval.
Data Availability
All data generated or analyzed during this study are included in this published article.
Abbreviations
cfDNA: cell-free DNA; cfDNA-mt: mitochondrial cell-free DNA; CpG: cytosine-phosphate-guanine; dd-cfDNA: donor-derived cell-free DNA; DSB: double-strand break; GF: gene transfer; HGT: horizontal gene transfer; HR: homologous recombination; NETs: neutrophil extracellular traps; NHEJ: non-homologous end joining; PRBCs: packed red blood cells; RBCs: red blood cells; TA-MC: transfusion-associated microchimerism; TLR: Toll-like receptor; WBCs: white blood cells
| References | ▴Top |
- Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, Samet Y, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068.
doi pubmed - Mattox AK, Douville C, Wang Y, Popoli M, Ptak J, Silliman N, Dobbyn L, et al. The origin of highly elevated cell-free dna in healthy individuals and patients with pancreatic, colorectal, lung, or ovarian cancer. Cancer Discov. 2023;13(10):2166-2179.
doi pubmed - Tsuji N, Agbor-Enoh S. Cell-free DNA beyond a biomarker for rejection: Biological trigger of tissue injury and potential therapeutics. J Heart Lung Transplant. 2021;40(6):405-413.
doi pubmed - Zukowski A, Rao S, Ramachandran S. Phenotypes from cell-free DNA. Open Biol. 2020;10(9):200119.
doi pubmed - Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1-2):57-68.
doi pubmed - Moldovan N, van der Pol Y, van den Ende T, Boers D, Verkuijlen S, Creemers A, Ramaker J, et al. Multi-modal cell-free DNA genomic and fragmentomic patterns enhance cancer survival and recurrence analysis. Cell Rep Med. 2024;5(1):101349.
doi pubmed - Stanley KE, Jatsenko T, Tuveri S, Sudhakaran D, Lannoo L, Van Calsteren K, de Borre M, et al. Cell type signatures in cell-free DNA fragmentation profiles reveal disease biology. Nat Commun. 2024;15(1):2220.
doi pubmed - Shi J, Zhang R, Li J, Zhang R. Size profile of cell-free DNA: A beacon guiding the practice and innovation of clinical testing. Theranostics. 2020;10(11):4737-4748.
doi pubmed - Lam WKJ, Gai W, Sun K, Wong RSM, Chan RWY, Jiang P, Chan NPH, et al. DNA of erythroid origin is present in human plasma and informs the types of anemia. Clin Chem. 2017;63(10):1614-1623.
doi pubmed - Wong FC, Sun K, Jiang P, Cheng YK, Chan KC, Leung TY, Chiu RW, et al. Cell-free DNA in maternal plasma and serum: A comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin Biochem. 2016;49(18):1379-1386.
doi pubmed - Pang Y, Andargie TE, Jang MK, Kong H, Park W, Hill T, Redekar N, et al. Chronic graft-versus-host disease is characterized by high levels and distinctive tissue-of-origin patterns of cell-free DNA. iScience. 2023;26(11):108160.
doi pubmed - Meszaros E. Understanding cell-free DNA: mechanisms, applications and analysis. Genomics. 2025.
- von Meijenfeldt FA, Burlage LC, Bos S, Adelmeijer J, Porte RJ, Lisman T. Elevated plasma levels of cell-free DNA during liver transplantation are associated with activation of coagulation. Liver Transpl. 2018;24(12):1716-1725.
doi pubmed - Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS One. 2017;12(8):e0183915.
doi pubmed - Bu J, Lee TH, Jeong WJ, Poellmann MJ, Mudd K, Eun HS, Liu EW, et al. Enhanced detection of cell-free DNA (cfDNA) enables its use as a reliable biomarker for diagnosis and prognosis of gastric cancer. PLoS One. 2020;15(12):e0242145.
doi pubmed - Yang L, Yang D, Yang Q, Cheng F, Huang Y. Extracellular DNA in blood products and its potential effects on transfusion. Biosci Rep. 2020;40(3):BSR20192770.
doi pubmed - Agbor-Enoh S, Chan JL, Singh A, Tunc I, Gorham S, Zhu J, Pirooznia M, et al. Circulating cell-free DNA as a biomarker of tissue injury: Assessment in a cardiac xenotransplantation model. J Heart Lung Transplant. 2018;37(8):967-975.
doi pubmed - Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18(2):134-147.
doi pubmed - Krenzien F, Katou S, Papa A, Sinn B, Benzing C, Feldbrugge L, Kamali C, et al. Increased cell-free DNA plasma concentration following liver transplantation is linked to portal hepatitis and inferior survival. J Clin Med. 2020;9(5):1543.
doi pubmed - Mallavia B, Liu F, Lefrancais E, Cleary SJ, Kwaan N, Tian JJ, Magnen M, et al. Mitochondrial DNA stimulates TLR9-dependent neutrophil extracellular trap formation in primary graft dysfunction. Am J Respir Cell Mol Biol. 2020;62(3):364-372.
doi pubmed - Liu Y, Peng F, Wang S, Jiao H, Zhou K, Guo W, Guo S, et al. Aberrant fragmentomic features of circulating cell-free mitochondrial DNA enable early detection and prognosis prediction of hepatocellular carcinoma. Clin Mol Hepatol. 2025;31(1):196-212.
doi pubmed - Zhu H, Wang Y, Li L, Wang L, Zhang H, Jin X. Cell-free DNA from clinical testing as a resource of population genetic analysis. Trends Genet. 2025;41(4):330-344.
doi pubmed - Ding B, Zhang X, Wan Z, Tian F, Ling J, Tan J, Peng X. Characterization of mitochondrial DNA methylation of Alzheimer's disease in plasma cell-free DNA. Diagnostics (Basel). 2023;13(14):2351.
doi pubmed - Hotz MJ, Qing D, Shashaty MGS, Zhang P, Faust H, Sondheimer N, Rivella S, et al. Red blood cells homeostatically bind mitochondrial DNA through TLR9 to maintain quiescence and to prevent lung injury. Am J Respir Crit Care Med. 2018;197(4):470-480.
doi pubmed - Lee CU, Cho E, Lee J, Lim JE, Chung JH, Song W, Kang M, et al. Chromosomal instability in cell-free DNA as a prognostic biomarker of metastatic hormone-sensitive prostate cancer treated with androgen deprivation therapy. Eur Urol Focus. 2023;9(1):89-95.
doi pubmed - Wong D, Tageldein M, Luo P, Ensminger E, Bruce J, Oldfield L, Gong H, et al. Cell-free DNA from germline TP53 mutation carriers reflect cancer-like fragmentation patterns. Nat Commun. 2024;15(1):7386.
doi pubmed - Doebley AL, Ko M, Liao H, Cruikshank AE, Santos K, Kikawa C, Hiatt JB, et al. A framework for clinical cancer subtyping from nucleosome profiling of cell-free DNA. Nat Commun. 2022;13(1):7475.
doi pubmed - Sivertsen J, Braathen H, Lunde THF, Kristoffersen EK, Hervig T, Strandenes G, Apelseth TO. Cold-stored leukoreduced CPDA-1 whole blood: in vitro quality and hemostatic properties. Transfusion. 2020;60(5):1042-1049.
doi pubmed - Nguyen PL, Asante A, Refaai M, Blumberg N. ABO blood group, ABO mismatched transfusions and leukoreduction of transfusions in hemostatic resuscitation studies. Front Bioeng Biotechnol. 2024;12:1437471.
doi pubmed - Waldvogel Abramowski S, Tirefort D, Lau P, Guichebaron A, Taleb S, Modoux C, Lemoine Chaduc C, et al. Cell-free nucleic acids are present in blood products and regulate genes of innate immune response. Transfusion. 2018;58(7):1671-1681.
doi pubmed - Pos O, Biro O, Szemes T, Nagy B. Circulating cell-free nucleic acids: characteristics and applications. Eur J Hum Genet. 2018;26(7):937-945.
doi pubmed - Waterhouse M, Themeli M, Bertz H, Zoumbos N, Finke J, Spyridonidis A. Horizontal DNA transfer from donor to host cells as an alternative mechanism of epithelial chimerism after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2011;17(3):319-329.
doi pubmed - Sender R, Noor E, Milo R, Dor Y. What fraction of cellular DNA turnover becomes cfDNA? Elife. 2024;12:RP89321.
doi pubmed - Jain V, Yang WH, Wu J, Roback JD, Gregory SG, Chi JT. Single cell RNA-Seq analysis of human red cells. Front Physiol. 2022;13:828700.
doi pubmed - Utter GH, Owings JT, Lee TH, Paglieroni TG, Reed WF, Gosselin RC, Holland PV, et al. Microchimerism in transfused trauma patients is associated with diminished donor-specific lymphocyte response. J Trauma. 2005;58(5):925-931; discussion 931-922.
doi pubmed - Gonzalez-Masia JA, Garcia-Olmo D, Garcia-Olmo DC. Circulating nucleic acids in plasma and serum (CNAPS): applications in oncology. Onco Targets Ther. 2013;6:819-832.
doi pubmed - Ktena YP, Dionysiou M, Gondek LP, Cooke KR. The impact of epigenetic modifications on allogeneic hematopoietic stem cell transplantation. Front Immunol. 2023;14:1188853.
doi pubmed - Zhang J, Li J, Saucier JB, Feng Y, Jiang Y, Sinson J, McCombs AK, et al. Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat Med. 2019;25(3):439-447.
doi pubmed - Sherwood K, Weimer ET. Characteristics, properties, and potential applications of circulating cell-free dna in clinical diagnostics: a focus on transplantation. J Immunol Methods. 2018;463:27-38.
doi pubmed - Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426-437.
doi pubmed - Sameer AS, Nissar S. Toll-Like Receptors (TLRs): structure, functions, signaling, and role of their polymorphisms in colorectal cancer susceptibility. Biomed Res Int. 2021;2021:1157023.
doi pubmed - Shrivastava S, Naik R, Suryawanshi H, Gupta N. Microchimerism: a new concept. J Oral Maxillofac Pathol. 2019;23(2):311.
doi pubmed - Matsagos S, Verigou E, Kourakli A, Alexis S, Vrakas S, Argyropoulou C, Lazaris V, et al. High frequency of post-transfusion microchimerism among multi-transfused beta-thalassemic patients. Front Med (Lausanne). 2022;9:845490.
doi pubmed - Malinska N, Grobarova V, Knizkova K, Cerny J. Maternal-Fetal Microchimerism: Impacts on Offspring's Immune Development and Transgenerational Immune Memory Transfer. Physiol Res. 2024;73(3):315-332.
doi pubmed - Kruchen A, Fehse B, Muller I. Clinical relevance of feto-maternal microchimerism in (hematopoietic stem cell) transplantation. Semin Immunopathol. 2024;47(1):4.
doi pubmed - Bloch EM, Busch MP, Lee TH, Montalvo L, Matthews Y, Bird A, Bruhn R, et al. Microchimerism in the transfused obstetric population. Vox Sang. 2014;107(4):428-430.
doi pubmed - Mo H, Wang X, Ma F, Qian Z, Sun X, Yi Z, Guan X, et al. Genome-wide chromosomal instability by cell-free DNA sequencing predicts survival in patients with metastatic breast cancer. Breast. 2020;53:111-118.
doi pubmed - Hirani R, Balogh ZJ, Lott NJ, Hsu JM, Irving DO. Leukodepleted blood components do not remove the potential for long-term transfusion-associated microchimerism in Australian major trauma patients. Chimerism. 2014;5(3-4):86-93.
doi pubmed - Benoit J, Leroy P, Vendrely R, Vendrely C. Experiments on Pekin ducks treated with DNA from Khaki Campbell ducks. Trans N Y Acad Sci. 1960;22:494-503.
doi pubmed - Puliyanda DP, Swinford R, Pizzo H, Garrison J, De Golovine AM, Jordan SC. Donor-derived cell-free DNA (dd-cfDNA) for detection of allograft rejection in pediatric kidney transplants. Pediatr Transplant. 2021;25(2):e13850.
doi pubmed - Aubert O, Ursule-Dufait C, Brousse R, Gueguen J, Racape M, Raynaud M, Van Loon E, et al. Cell-free DNA for the detection of kidney allograft rejection. Nat Med. 2024;30(8):2320-2327.
doi pubmed - McCoy CC, Brenner M, Duchesne J, Roberts D, Ferrada P, Horer T, Kauvar D, et al. Back to the Future: Whole Blood Resuscitation of the Severely Injured Trauma Patient. Shock. 2021;56(1S):9-15.
doi pubmed - Osataphan N, Dumnil S, Tantiworawit A, Punnachet T, Hantrakun N, Piriyakhuntorn P, Rattanathammethee T, et al. The long-term efficacy in blood transfusions, hematologic parameter changes, and complications after splenectomy in patients with transfusion-dependent thalassemia. Transfus Apher Sci. 2023;62(3):103620.
doi pubmed - Pisareva E, Mihalovicova L, Pastor B, Kudriavtsev A, Mirandola A, Mazard T, Badiou S, et al. Neutrophil extracellular traps have auto-catabolic activity and produce mononucleosome-associated circulating DNA. Genome Med. 2022;14(1):135.
doi pubmed - Shaik-Dasthagirisaheb YB, Varvara G, Murmura G, Saggini A, Potalivo G, Caraffa A, Antinolfi P, et al. Vascular endothelial growth factor (VEGF), mast cells and inflammation. Int J Immunopathol Pharmacol. 2013;26(2):327-335.
doi pubmed - Lukic A, Larssen P, Fauland A, Samuelsson B, Wheelock CE, Gabrielsson S, Radmark O. GM-CSF- and M-CSF-primed macrophages present similar resolving but distinct inflammatory lipid mediator signatures. FASEB J. 2017;31(10):4370-4381.
doi pubmed - Chatterjee M, Rauen T, Kis-Toth K, Kyttaris VC, Hedrich CM, Terhorst C, Tsokos GC. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol. 2012;188(3):1206-1212.
doi pubmed - Chan RW, Jiang P, Peng X, Tam LS, Liao GJ, Li EK, Wong PC, et al. Plasma DNA aberrations in systemic lupus erythematosus revealed by genomic and methylomic sequencing. Proc Natl Acad Sci U S A. 2014;111(49):E5302-5311.
doi pubmed - Wang Y, Zhu J, Zhang L, Zhang Z, He L, Mou Y, Deng Y, et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor alpha positive feedback loop in M2 macrophages. J Allergy Clin Immunol. 2017;140(6):1550-1561.e1558.
doi pubmed - Tsuji N, Tsuji T, Ohashi N, Kato A, Fujigaki Y, Yasuda H. Role of mitochondrial DNA in septic AKI via toll-like receptor 9. J Am Soc Nephrol. 2016;27(7):2009-2020.
doi pubmed - Lorenzo-Diaz F, Fernandez-Lopez C, Lurz R, Bravo A, Espinosa M. Crosstalk between vertical and horizontal gene transfer: plasmid replication control by a conjugative relaxase. Nucleic Acids Res. 2017;45(13):7774-7785.
doi pubmed - Berger B, Parson R, Clausen J, Berger C, Nachbaur D, Parson W. Chimerism in DNA of buccal swabs from recipients after allogeneic hematopoietic stem cell transplantations: implications for forensic DNA testing. Int J Legal Med. 2013;127(1):49-54.
doi pubmed - Garcia-Olmo DC, Dominguez C, Garcia-Arranz M, Anker P, Stroun M, Garcia-Verdugo JM, Garcia-Olmo D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010;70(2):560-567.
doi pubmed - Olivera-Salazar R, Garcia-Arranz M, Sanchez A, Olmedillas-Lopez S, Vega-Clemente L, Serrano LJ, Herrera B, et al. Oncological transformation in vitro of hepatic progenitor cell lines isolated from adult mice. Sci Rep. 2022;12(1):3149.
doi pubmed - Al Amir Dache Z, Thierry AR. Mitochondria-derived cell-to-cell communication. Cell Rep. 2023;42(7):112728.
doi pubmed - Holmgren L, Szeles A, Rajnavolgyi E, Folkman J, Klein G, Ernberg I, Falk KI. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood. 1999;93(11):3956-3963.
pubmed - Zuo Y, Zuo M, Yalavarthi S, Gockman K, Madison JA, Shi H, Woodard W, et al. Neutrophil extracellular traps and thrombosis in COVID-19. J Thromb Thrombolysis. 2021;51(2):446-453.
doi pubmed - Korabecna M, Zinkova A, Brynychova I, Chylikova B, Prikryl P, Sedova L, Neuzil P, et al. Cell-free DNA in plasma as an essential immune system regulator. Sci Rep. 2020;10(1):17478.
doi pubmed - Sayah DM, Mallavia B, Liu F, Ortiz-Munoz G, Caudrillier A, DerHovanessian A, Ross DJ, et al. Neutrophil extracellular traps are pathogenic in primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2015;191(4):455-463.
doi pubmed - Mondelo-Macia P, Castro-Santos P, Castillo-Garcia A, Muinelo-Romay L, Diaz-Pena R. Circulating free DNA and its emerging role in autoimmune diseases. J Pers Med. 2021;11(2):151.
doi pubmed - Charoensappakit A, Sae-Khow K, Rattanaliam P, Vutthikraivit N, Pecheenbuvan M, Udomkarnjananun S, Leelahavanichkul A. Cell-free DNA as diagnostic and prognostic biomarkers for adult sepsis: a systematic review and meta-analysis. Sci Rep. 2023;13(1):19624.
doi pubmed - Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, Quintana C, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10(12):e1001577; discussion e1001577.
doi pubmed - Pietrzak B, Kawacka I, Olejnik-Schmidt A, Schmidt M. Circulating microbial cell-free DNA in health and disease. Int J Mol Sci. 2023;24(3):3051.
doi pubmed - Aydin S, Ozdemir S, Adiguzel A. The potential of cfDNA as biomarker: opportunities and challenges for neurodegenerative diseases. J Mol Neurosci. 2025;75(1):34.
doi pubmed - Melamud MM, Buneva VN, Ermakov EA. Circulating cell-free DNA levels in psychiatric diseases: a systematic review and meta-analysis. Int J Mol Sci. 2023;24(4):3402.
doi pubmed - McSorley ST, Tham A, Dolan RD, Steele CW, Ramsingh J, Roxburgh C, Horgan PG, et al. Perioperative blood transfusion is associated with postoperative systemic inflammatory response and poorer outcomes following surgery for colorectal cancer. Ann Surg Oncol. 2020;27(3):833-843.
doi pubmed - Lopes MA, Cordeiro MER, de Alencar Teles Barreto F, de Souza Moreno L, de Medeiros Silva AA, de Loyola MB, Soares MVA, et al. Assessment of cfDNA release dynamics during colorectal cancer surgery. Oncotarget. 2025;16:29-38.
doi pubmed - Zhang W, Jing X, Li B, Wu X. Clearance of cell-free DNA: a novel target for therapeutic utilization in multiple systemic disorders. ACS Biomater Sci Eng. 2025;11(4):2069-2079.
doi pubmed - Bronkhorst AJ, Ungerer V, Oberhofer A, Gabriel S, Polatoglou E, Randeu H, Uhlig C, et al. New perspectives on the importance of cell-free DNA biology. Diagnostics (Basel). 2022;12(9):2147.
doi pubmed - Jun YW, Kant M, Coskun E, Kato TA, Jaruga P, Palafox E, Dizdaroglu M, et al. Possible genetic risks from heat-damaged DNA in food. ACS Cent Sci. 2023;9(6):1170-1179.
doi pubmed - Schubbert R, Hohlweg U, Renz D, Doerfler W. On the fate of orally ingested foreign DNA in mice: chromosomal association and placental transmission to the fetus. Mol Gen Genet. 1998;259(6):569-576.
doi pubmed - Spisak S, Solymosi N, Ittzes P, Bodor A, Kondor D, Vattay G, Bartak BK, et al. Complete genes may pass from food to human blood. PLoS One. 2013;8(7):e69805.
doi pubmed - Lerner A, Benzvi C, Vojdani A. The potential harmful effects of genetically engineered microorganisms (GEMs) on the intestinal microbiome and public health. Microorganisms. 2024;12(2):238.
doi pubmed - Yang Z, Zeng J, Chen Y, Wang M, Luo H, Huang AL, Deng H, et al. Detection of HBV DNA integration in plasma cell-free DNA of different HBV diseases utilizing DNA capture strategy. Virol Sin. 2024;39(4):655-666.
doi pubmed - Fritsche S, Reinfurt A, Fronek F, Steiger MG. NHEJ and HDR can occur simultaneously during gene integration into the genome of Aspergillus niger. Fungal Biol Biotechnol. 2024;11(1):10.
doi pubmed - Liester MB. Personality changes following heart transplantation: the role of cellular memory. Med Hypotheses. 2020;135:109468.
doi pubmed - Dvorakova M, Karafiat V, Pajer P, Kluzakova E, Jarkovska K, Pekova S, Krutilkova L, et al. DNA released by leukemic cells contributes to the disruption of the bone marrow microenvironment. Oncogene. 2013;32(44):5201-5209.
doi pubmed - Pearsall P, Schwartz GE, Russek LG. Changes in heart transplant recipients that parallel the personalities of their donors. Integr Med. 2000;2(2):65-72.
doi pubmed - Allen L, Dwivedi Y. MicroRNA mediators of early life stress vulnerability to depression and suicidal behavior. Mol Psychiatry. 2020;25(2):308-320.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.